[轉]常見醫療掃描影象處理步驟
一 資料格式
1.1 dicom
DICOM是醫學影象中標準檔案,這些檔案包含了諸多的元資料資訊(比如畫素尺寸,每個維度的一畫素代表真實世界裡的長度)。此處以kaggle Data Science Bowl 資料集為例。
data-science-bowl-2017。資料列表如下:
字尾為 .dcm。
每個病人的一次掃描CT(scan)可能有幾十到一百多個dcm資料檔案(slices)。可以使用 python的dicom包讀取,讀取示例程式碼如下:
dicom.read_file('/data/lung_competition/stage1/7050f8141e92fa42fd9c471a8b2f50ce/498d16aa2222d76cae1da144ddc59a13.dcm')
其pixl_array包含了真實資料。
slices = [dicom.read_file(os.path.join(folder_name,filename)) for filename in os.listdir(folder_name)]slices = np.stack([s.pixel_array for s in slices])
1.2 mhd格式
mhd格式是另外一種資料格式,來源於(LUNA2016)[https://luna16.grand-challenge.org/data/]。每個病人一個mhd檔案和一個同名的raw
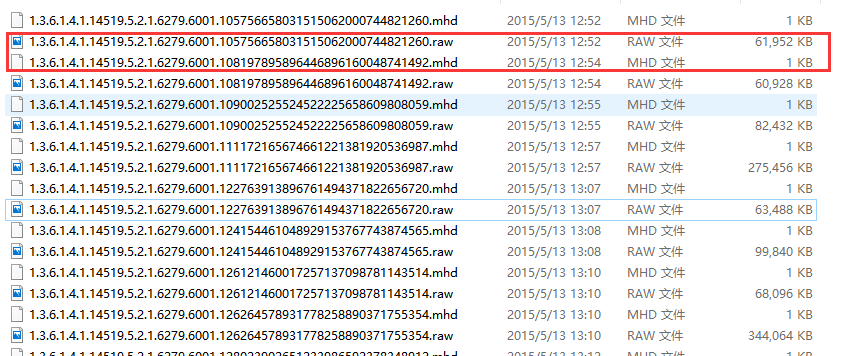
一個mhd通常有幾百兆,對應的raw檔案只有1kb。mhd檔案需要藉助python的SimpleITK包來處理。SimpleITK 示例程式碼如下:
import SimpleITK as sitkitk_img = sitk.ReadImage(img_file)img_array = sitk.GetArrayFromImage(itk_img) # indexes are z,y,x (notice the ordering)num_z, height, width = img_array.shape #heightXwidth constitute the transverse planeorigin = np.array(itk_img.GetOrigin()) # x,y,z Origin in world coordinates (mm)spacing = np.array(itk_img.GetSpacing()) # spacing of voxels in world coor. (mm)
需要注意的是,SimpleITK的img_array的陣列不是直接的畫素值,而是相對於CT掃描中原點位置的差值,需要做進一步轉換。轉換步驟參考 SimpleITK影象轉換
1.3 檢視CT掃描檔案軟體
一個開源免費的檢視軟體 mango
二 dicom格式資料處理過程
2.1 處理思路
首先,需要明白的是醫學掃描影象(scan)其實是三維影象,使用程式碼讀取之後開源檢視不同的切面的切片(slices),可以從不同軸切割
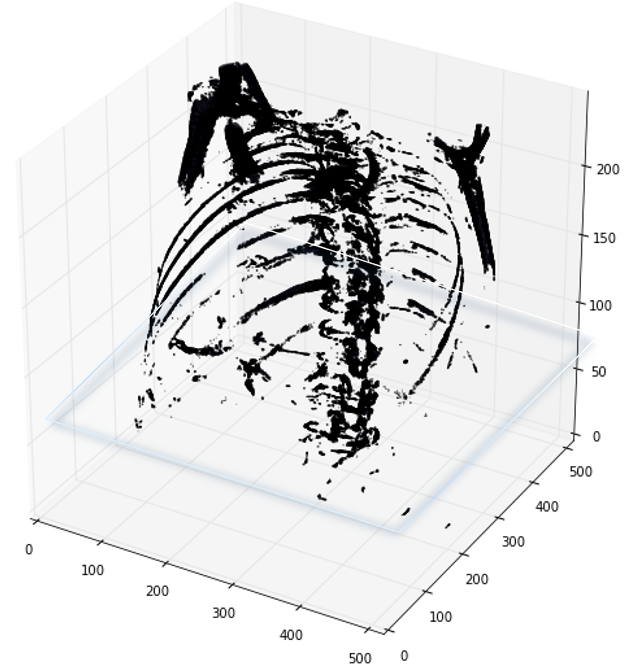
如下圖展示了一個病人CT掃描圖中,其中部分切片slices
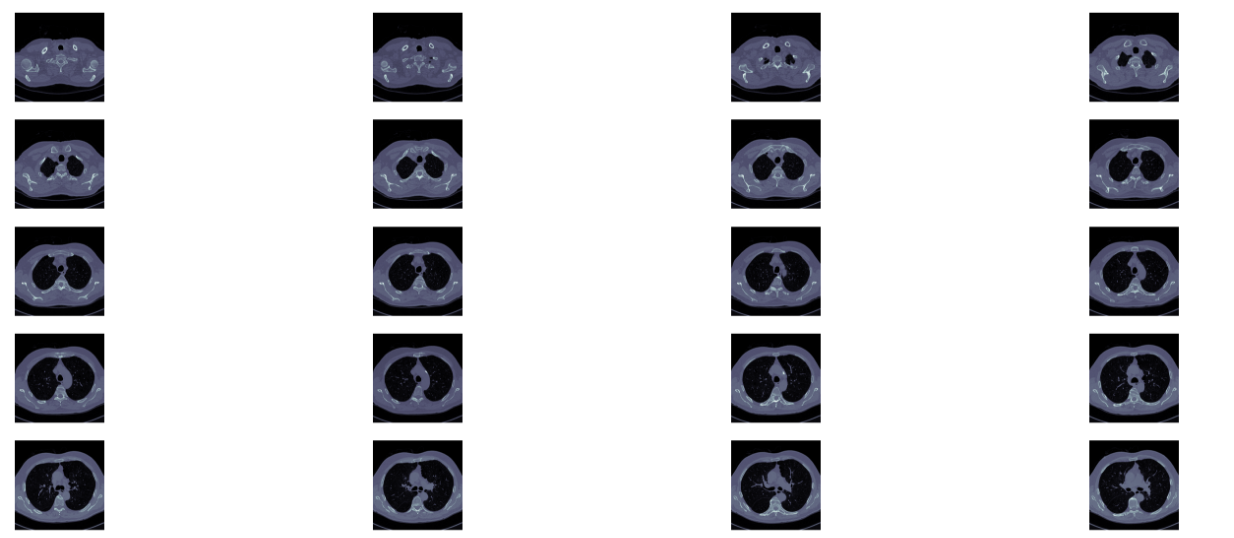
其次,CT掃描圖是包含了所有組織的,如果直接去看,看不到任何有用資訊。需要做一些預處理,預處理中一個重要的概念是放射劑量,衡量單位為HU(Hounsfield Unit),下表是不同放射劑量對應的組織器官
| substance | HU |
|---|---|
| 空氣 | -1000 |
| 肺 | -500 |
| 脂肪 | -100到-50 |
| 水 | 0 |
| CSF | 15 |
| 腎 | 30 |
| 血液 | +30到+45 |
| 肌肉 | +10到+40 |
| 灰質 | +37到+45 |
| 白質 | +20到+30 |
| Liver | +40到+60 |
| 軟組織,constrast | +100到+300 |
| 骨頭 | +700(軟質骨)到+3000(皮質骨) |
Hounsfield Unit = pixel_value * rescale_slope + rescale_intercept
一般情況rescale slope = 1, intercept = -1024。
上表中肺部組織的HU數值為-500,但通常是大於這個值,比如-320、-400。挑選出這些區域,然後做其他變換抽取出肺部畫素點。
2.2 先載入必要的包
# -*- coding:utf-8 -*-'''this script is used for basic process of lung 2017 in Data Science Bowl'''import globimport osimport pandas as pdimport SimpleITK as sitkimport numpy as np # linear algebraimport pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)import skimage, osfrom skimage.morphology import ball, disk, dilation, binary_erosion, remove_small_objects, erosion, closing, reconstruction, binary_closingfrom skimage.measure import label,regionprops, perimeterfrom skimage.morphology import binary_dilation, binary_openingfrom skimage.filters import roberts, sobelfrom skimage import measure, featurefrom skimage.segmentation import clear_borderfrom skimage import datafrom scipy import ndimage as ndiimport matplotlib#matplotlib.use('Agg')import matplotlib.pyplot as pltfrom mpl_toolkits.mplot3d.art3d import Poly3DCollectionimport dicomimport scipy.miscimport numpy as np
如下程式碼是載入一個掃描面,包含了多個切片(slices),我們僅簡化的將其儲存為python列表。資料集中每個目錄都是一個掃描面集(一個病人)。有個元資料域丟失,即Z軸方向上的畫素尺寸,也即切片的厚度 。所幸,我們可以用其他值推測出來,並加入到元資料中。
# Load the scans in given folder pathdef load_scan(path):slices = [dicom.read_file(path + '/' + s) for s in os.listdir(path)]slices.sort(key = lambda x: int(x.ImagePositionPatient[2]))try:slice_thickness = np.abs(slices[0].ImagePositionPatient[2] - slices[1].ImagePositionPatient[2])except:slice_thickness = np.abs(slices[0].SliceLocation - slices[1].SliceLocation)for s in slices:s.SliceThickness = slice_thicknessreturn slices
2.3 灰度值轉換為HU單元
首先去除灰度值為 -2000的pixl_array,CT掃描邊界之外的灰度值固定為-2000(dicom和mhd都是這個值)。第一步是設定這些值為0,當前對應為空氣(值為0)
回到HU單元,乘以rescale比率並加上intercept(儲存在掃描面的元資料中)。
def get_pixels_hu(slices):image = np.stack([s.pixel_array for s in slices])# Convert to int16 (from sometimes int16),# should be possible as values should always be low enough (<32k)image = image.astype(np.int16)# Set outside-of-scan pixels to 0# The intercept is usually -1024, so air is approximately 0image[image == -2000] = 0# Convert to Hounsfield units (HU)for slice_number in range(len(slices)):intercept = slices[slice_number].RescaleInterceptslope = slices[slice_number].RescaleSlopeif slope != 1:image[slice_number] = slope * image[slice_number].astype(np.float64)image[slice_number] = image[slice_number].astype(np.int16)image[slice_number] += np.int16(intercept)return np.array(image, dtype=np.int16)
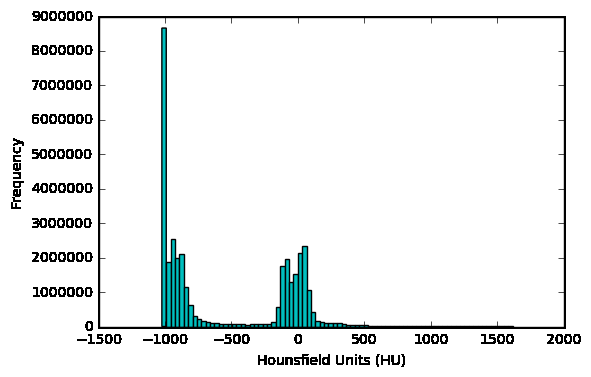
可以檢視病人的掃描HU值分佈情況
first_patient = load_scan(INPUT_FOLDER + patients[0])first_patient_pixels = get_pixels_hu(first_patient)plt.hist(first_patient_pixels.flatten(), bins=80, color='c')plt.xlabel("Hounsfield Units (HU)")plt.ylabel("Frequency")plt.show()
2.4 重取樣
不同掃描面的畫素尺寸、粗細粒度是不同的。這不利於我們進行CNN任務,我們可以使用同構取樣。
一個掃描面的畫素區間可能是[2.5,0.5,0.5],即切片之間的距離為2.5mm。可能另外一個掃描面的範圍是[1.5,0.725,0.725]。這可能不利於自動分析。
常見的處理方法是從全資料集中以固定的同構解析度重新取樣,將所有的東西取樣為1mmx1mmx1mm畫素。
def resample(image, scan, new_spacing=[1,1,1]):# Determine current pixel spacingspacing = map(float, ([scan[0].SliceThickness] + scan[0].PixelSpacing))spacing = np.array(list(spacing))resize_factor = spacing / new_spacingnew_real_shape = image.shape * resize_factornew_shape = np.round(new_real_shape)real_resize_factor = new_shape / image.shapenew_spacing = spacing / real_resize_factorimage = scipy.ndimage.interpolation.zoom(image, real_resize_factor, mode='nearest')return image, new_spacing
現在重新取樣病人的畫素,將其對映到一個同構解析度 1mm x1mm x1mm。
pix_resampled, spacing = resample(first_patient_pixels, first_patient, [1,1,1])
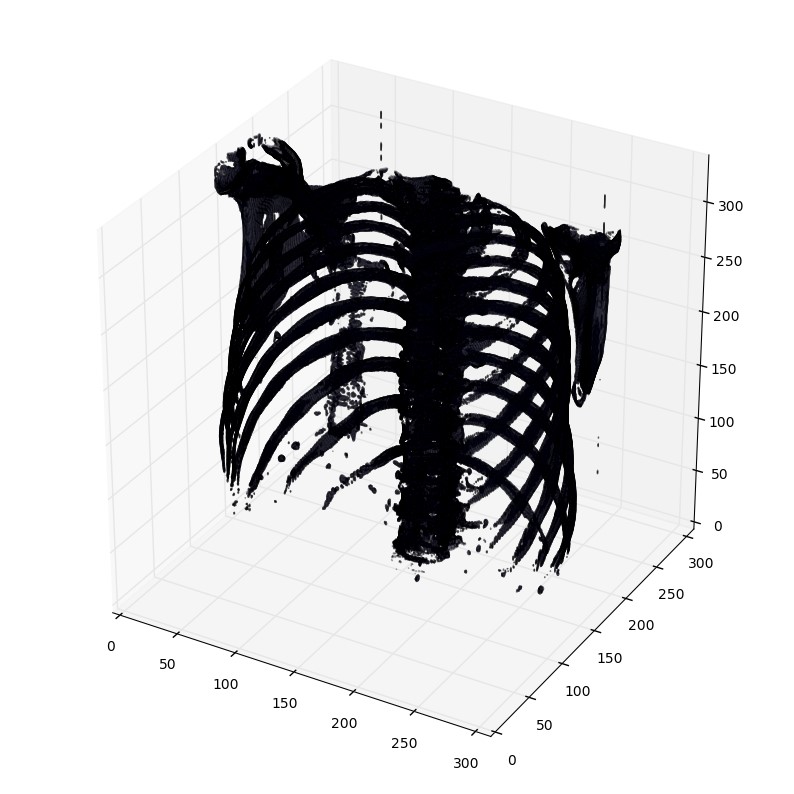
使用matplotlib輸出肺部掃描的3D影象方法。可能需要一兩分鐘。
def plot_3d(image, threshold=-300):# Position the scan upright,# so the head of the patient would be at the top facing the camerap = image.transpose(2,1,0)verts, faces = measure.marching_cubes(p, threshold)fig = plt.figure(figsize=(10, 10))ax = fig.add_subplot(111, projection='3d')# Fancy indexing: `verts[faces]` to generate a collection of trianglesmesh = Poly3DCollection(verts[faces], alpha=0.1)face_color = [0.5, 0.5, 1]mesh.set_facecolor(face_color)ax.add_collection3d(mesh)ax.set_xlim(0, p.shape[0])ax.set_ylim(0, p.shape[1])ax.set_zlim(0, p.shape[2])plt.show()
列印函式有個閾值引數,來列印特定的結構,比如tissue或者骨頭。400是一個僅僅列印骨頭的閾值(HU對照表)
plot_3d(pix_resampled, 400)
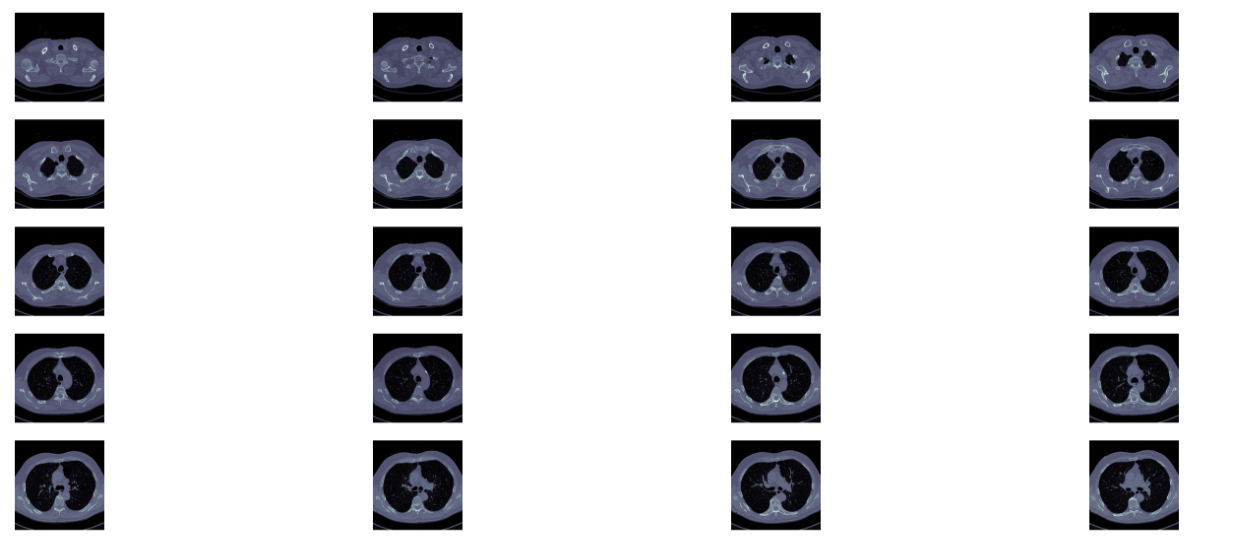
2.5 輸出一個病人scans中所有切面slices
def plot_ct_scan(scan):'''plot a few more images of the slices:param scan::return:'''f, plots = plt.subplots(int(scan.shape[0] / 20) + 1, 4, figsize=(50, 50))for i in range(0, scan.shape[0], 5):plots[int(i / 20), int((i % 20) / 5)].axis('off')plots[int(i / 20), int((i % 20) / 5)].imshow(scan[i], cmap=plt.cm.bone)
此方法的效果示例如下:
2.6 定義分割出CT切面裡面肺部組織的函式
下面的程式碼使用了pythonde 的影象形態學操作。具體可以參考python高階形態學操作
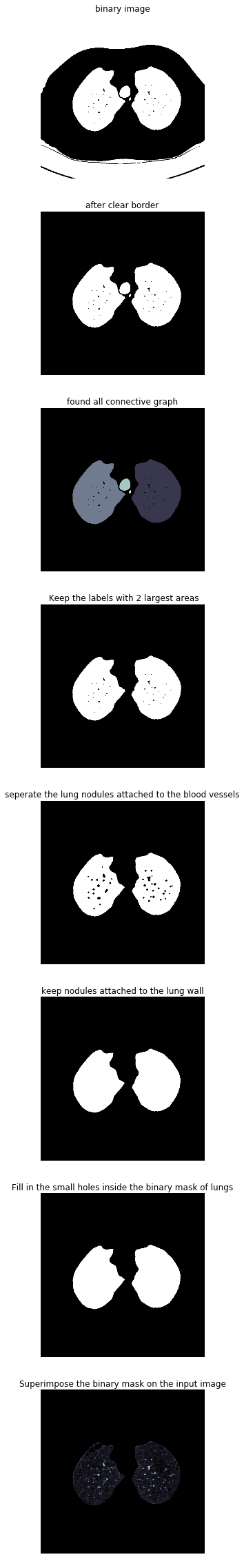
def get_segmented_lungs(im, plot=False):'''This funtion segments the lungs from the given 2D slice.'''if plot == True:f, plots = plt.subplots(8, 1, figsize=(5, 40))'''Step 1: Convert into a binary image.'''binary = im < 604if plot == True:plots[0].axis('off')plots[0].set_title('binary image')plots[0].imshow(binary, cmap=plt.cm.bone)'''Step 2: Remove the blobs connected to the border of the image.'''cleared = clear_border(binary)if plot == True:plots[1].axis('off')plots[1].set_title('after clear border')plots[1].imshow(cleared, cmap=plt.cm.bone)'''Step 3: Label the image.'''label_image = label(cleared)if plot == True:plots[2].axis('off')plots[2].set_title('found all connective graph')plots[2].imshow(label_image, cmap=plt.cm.bone)'''Step 4: Keep the labels with 2 largest areas.'''areas = [r.area for r in regionprops(label_image)]areas.sort()if len(areas) > 2:for region in regionprops(label_image):if region.area < areas[-2]:for coordinates in region.coords:label_image[coordinates[0], coordinates[1]] = 0binary = label_image > 0if plot == True:plots[3].axis('off')plots[3].set_title(' Keep the labels with 2 largest areas')plots[3].imshow(binary, cmap=plt.cm.bone)'''Step 5: Erosion operation with a disk of radius 2. This operation isseperate the lung nodules attached to the blood vessels.'''selem = disk(2)binary = binary_erosion(binary, selem)if plot == True:plots[4].axis('off')plots[4].set_title('seperate the lung nodules attached to the blood vessels')plots[4].imshow(binary, cmap=plt.cm.bone)'''Step 6: Closure operation with a disk of radius 10. This operation isto keep nodules attached to the lung wall.'''selem = disk(10)binary = binary_closing(binary, selem)if plot == True:plots[5].axis('off')plots[5].set_title('keep nodules attached to the lung wall')plots[5].imshow(binary, cmap=plt.cm.bone)'''Step 7: Fill in the small holes inside the binary mask of lungs.'''edges = roberts(binary)binary = ndi.binary_fill_holes(edges)if plot == True:plots[6].axis('off')plots[6].set_title('Fill in the small holes inside the binary mask of lungs')plots[6].imshow(binary, cmap=plt.cm.bone)'''Step 8: Superimpose the binary mask on the input image.'''get_high_vals = binary == 0im[get_high_vals] = 0if plot == True:plots[7].axis('off')plots[7].set_title('Superimpose the binary mask on the input image')plots[7].imshow(im, cmap=plt.cm.bone)return im
此方法每個步驟對影象做不同的處理,依次為二值化、清除邊界、連通區域標記、腐蝕操作、閉合運算、孔洞填充、效果如下:
2.7 肺部影象分割
為了減少有問題的空間,我們可以分割肺部影象(有時候是附近的組織)。這包含一些步驟,包括區域增長和形態運算,此時,我們只分析相連元件。
步驟如下:
-
閾值影象(-320HU是個極佳的閾值,但是此方法中不是必要)
-
處理相連的元件,以決定當前患者的空氣的標籤,以1填充這些二值影象
-
可選:當前掃描的每個軸上的切片,選定最大固態連線的組織(當前患者的肉體和空氣),並且其他的為0。以掩碼的方式填充肺部結構。
-
只保留最大的氣袋(人類軀體內到處都有氣袋)
def largest_label_volume(im, bg=-1):vals, counts = np.unique(im, return_counts=True)counts = counts[vals != bg]vals = vals[vals != bg]if len(counts) > 0:return vals[np.argmax(counts)]else:return Nonedef segment_lung_mask(image, fill_lung_structures=True):# not actually binary, but 1 and 2.# 0 is treated as background, which we do not wantbinary_image = np.array(image > -320, dtype=np.int8)+1labels = measure.label(binary_image)
