用python實現簡單的遺傳演算法
阿新 • • 發佈:2019-01-24
首先遺傳演算法是一種優化演算法,通過模擬基因的優勝劣汰,進行計算(具體的演算法思路什麼的就不贅述了)。大致過程分為初始化編碼、個體評價、選擇,交叉,變異。
以目標式子 y = 10 * sin(5x) + 7 * cos(4x)為例,計算其最大值
首先是初始化,包括具體要計算的式子、種群數量、染色體長度、交配概率、變異概率等。並且要對基因序列進行初始化
- pop_size = 500# 種群數量
- max_value = 10# 基因中允許出現的最大值
- chrom_length = 10# 染色體長度
- pc = 0.6# 交配概率
- pm = 0.01# 變異概率
- results = [[]] # 儲存每一代的最優解,N個二元組
- fit_value = [] # 個體適應度
- fit_mean = [] # 平均適應度
- pop = geneEncoding(pop_size, chrom_length)
其中genEncodeing是自定義的一個簡單隨機生成序列的函式,具體實現如下
- def geneEncoding(pop_size, chrom_length):
- pop = [[]]
- for i in range(pop_size):
- temp = []
- for j in range(chrom_length):
- temp.append(random.randint(0, 1))
- pop.append(temp)
- return pop[1:]
編碼完成之後就是要進行個體評價,個體評價主要是計算各個編碼出來的list的值以及對應帶入目標式子的值。其實編碼出來的就是一堆2進位制list。這些2進位制list每個都代表了一個數。其值的計算方式為轉換為10進位制,然後除以2的序列長度次方減一,也就是全一list的十進位制減一。根據這個規則就能計算出所有list的值和帶入要計算式子中的值,程式碼如下
- # 0.0 coding:utf-8 0.0
- # 解碼並計算值
- import math
- def decodechrom(pop, chrom_length):
- temp = []
- for i in range(len(pop)):
- t = 0
- for j in range(chrom_length):
- t += pop[i][j] * (math.pow(2, j))
- temp.append(t)
- return temp
- def calobjValue(pop, chrom_length, max_value):
- temp1 = []
- obj_value = []
- temp1 = decodechrom(pop, chrom_length)
- for i in range(len(temp1)):
- x = temp1[i] * max_value / (math.pow(2, chrom_length) - 1)
- obj_value.append(10 * math.sin(5 * x) + 7 * math.cos(4 * x))
- return obj_value
有了具體的值和對應的基因序列,然後進行一次淘汰,目的是淘汰掉一些不可能的壞值。這裡由於是計算最大值,於是就淘汰負值就好了
- # 0.0 coding:utf-8 0.0
- # 淘汰(去除負值)
- def calfitValue(obj_value):
- fit_value = []
- c_min = 0
- for i in range(len(obj_value)):
- if(obj_value[i] + c_min > 0):
- temp = c_min + obj_value[i]
- else:
- temp = 0.0
- fit_value.append(temp)
- return fit_value
然後就是進行選擇,這是整個遺傳演算法最核心的部分。選擇實際上模擬生物遺傳進化的優勝劣汰,讓優秀的個體儘可能存活,讓差的個體儘可能的淘汰。個體的好壞是取決於個體適應度。個體適應度越高,越容易被留下,個體適應度越低越容易被淘汰。具體的程式碼如下
- # 0.0 coding:utf-8 0.0
- # 選擇
- import random
- def sum(fit_value):
- total = 0
- for i in range(len(fit_value)):
- total += fit_value[i]
- return total
- def cumsum(fit_value):
- for i in range(len(fit_value)-2, -1, -1):
- t = 0
- j = 0
- while(j <= i):
- t += fit_value[j]
- j += 1
- fit_value[i] = t
- fit_value[len(fit_value)-1] = 1
- def selection(pop, fit_value):
- newfit_value = []
- # 適應度總和
- total_fit = sum(fit_value)
- for i in range(len(fit_value)):
- newfit_value.append(fit_value[i] / total_fit)
- # 計算累計概率
- cumsum(newfit_value)
- ms = []
- pop_len = len(pop)
- for i in range(pop_len):
- ms.append(random.random())
- ms.sort()
- fitin = 0
- newin = 0
- newpop = pop
- # 轉輪盤選擇法
- while newin < pop_len:
- if(ms[newin] < newfit_value[fitin]):
- newpop[newin] = pop[fitin]
- newin = newin + 1
- else:
- fitin = fitin + 1
- pop = newpop
選擇完後就是進行交配和變異,這個兩個步驟很好理解。就是對基因序列進行改變,只不過改變的方式不一樣
交配:
- # 0.0 coding:utf-8 0.0
- # 交配
- import random
- def crossover(pop, pc):
- pop_len = len(pop)
- for i in range(pop_len - 1):
- if(random.random() < pc):
- cpoint = random.randint(0,len(pop[0]))
- temp1 = []
- temp2 = []
- temp1.extend(pop[i][0:cpoint])
- temp1.extend(pop[i+1][cpoint:len(pop[i])])
- temp2.extend(pop[i+1][0:cpoint])
- temp2.extend(pop[i][cpoint:len(pop[i])])
- pop[i] = temp1
- pop[i+1] = temp2
變異:
- # 0.0 coding:utf-8 0.0
- # 基因突變
- import random
- def mutation(pop, pm):
- px = len(pop)
- py = len(pop[0])
- for i in range(px):
- if(random.random() < pm):
- mpoint = random.randint(0, py-1)
- if(pop[i][mpoint] == 1):
- pop[i][mpoint] = 0
- else:
- pop[i][mpoint] = 1
整個遺傳演算法的實現完成了,總的呼叫入口程式碼如下
- # 0.0 coding:utf-8 0.0
- import matplotlib.pyplot as plt
- import math
- from calobjValue import calobjValue
- from calfitValue import calfitValue
- from selection import selection
- from crossover import crossover
- from mutation import mutation
- from best import best
- from geneEncoding import geneEncoding
- print'y = 10 * math.sin(5 * x) + 7 * math.cos(4 * x)'
- # 計算2進位制序列代表的數值
- def b2d(b, max_value, chrom_length):
- t = 0
- for j in range(len(b)):
- t += b[j] * (math.pow(2, j))
- t = t * max_value / (math.pow(2, chrom_length) - 1)
- return t
- pop_size = 500# 種群數量
- max_value = 10# 基因中允許出現的最大值
- chrom_length = 10# 染色體長度
- pc = 0.6# 交配概率
- pm = 0.01# 變異概率
- results = [[]] # 儲存每一代的最優解,N個二元組
- fit_value = [] # 個體適應度
- fit_mean = [] # 平均適應度
- # pop = [[0, 1, 0, 1, 0, 1, 0, 1, 0, 1] for i in range(pop_size)]
- pop = geneEncoding(pop_size, chrom_length)
- for i in range(pop_size):
- obj_value = calobjValue(pop, chrom_length, max_value) # 個體評價
- fit_value = calfitValue(obj_value) # 淘汰
- best_individual, best_fit = best(pop, fit_value) # 第一個儲存最優的解, 第二個儲存最優基因
- results.append([best_fit, b2d(best_individual, max_value, chrom_length)])
- selection(pop, fit_value) # 新種群複製
- crossover(pop, pc) # 交配
- mutation(pop, pm) # 變異
- results = results[1:]
- results.sort()
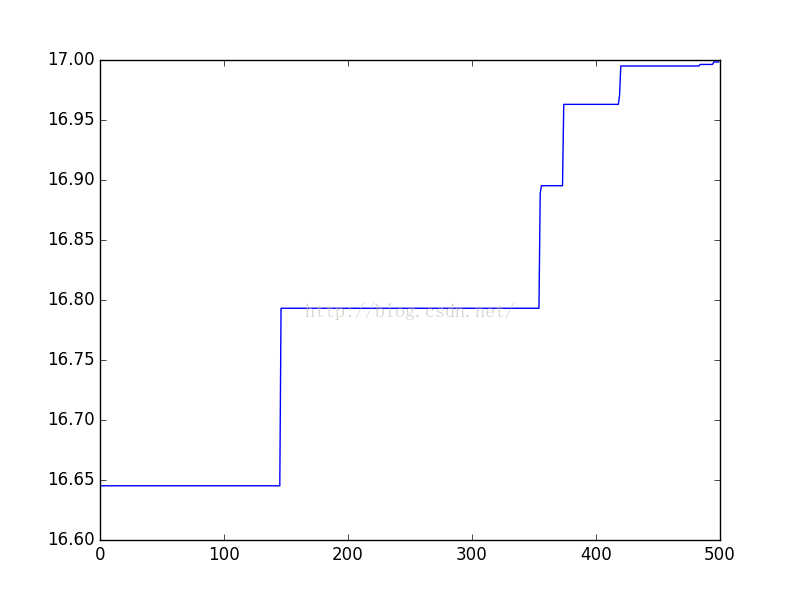
- X = []
- Y = []
- for i in range(500):
- X.append(i)
- t = results[i][0]
- Y.append(t)
- plt.plot(X, Y)
- plt.show()
完整程式碼可以在github 檢視