機器學習-多分類問題
一、背景
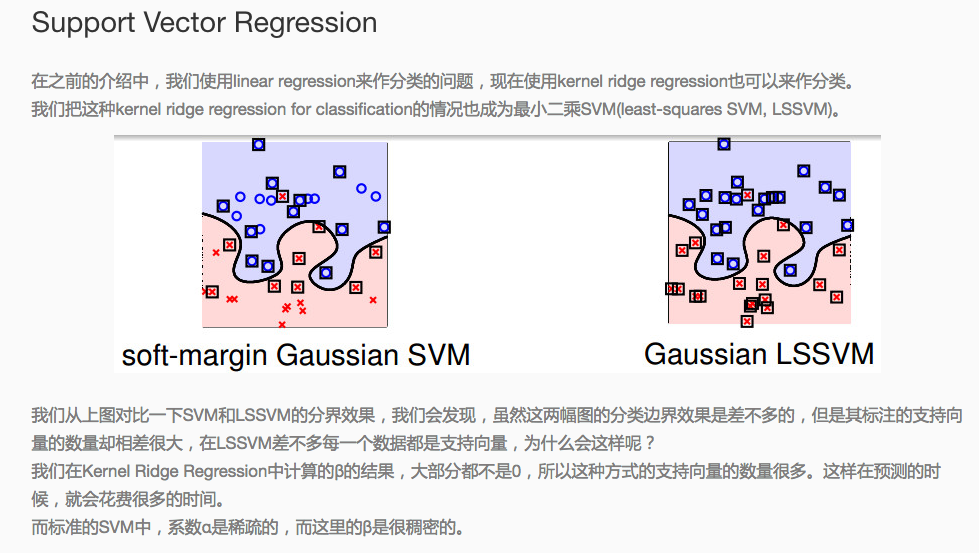
為何選擇svm來嘗試解決多分類問題,以下為決策樹和svm用於多分類的表現上的差異:
二、SVM支援向量機(Support vector machine)
- 設計k個SVM兩類分類器;
- 設計兩兩k(k-1)/2個SVM兩類分類器。
- 線上性方程後加高階項:採用一次優化求解解決問題。對於每一類,設計w_i與b_i,約束真實類別對應的w_i x + b_i大於其他類別的w_i x + b_i進行訓練,求解目標是所有w_i的範數之和最小,也可以引入 樣本數乘以類別數 個鬆馳變數
三、SVR支援向量迴歸(Support vector Regression)
原理:用核函式代替線性方程中的線性項可以使原來的線性演算法“非線性化”,即能做非線性迴歸。引進核函式達到了“升維”的目的,而增加的可調引數使得過擬合依然能控制。
- 所謂迴歸(regression),基本上就是擬合,用一個函式擬合x與y的關係。對於SVR來說,x是向量,y是標量,擬合的函式形式為y=W^T*g(x)+b,其中g(x)為核函式對應的特徵空間向量。
- SVR認為,只要估計的y在實際的y的兩側一個固定的範圍(epsilon)之內,就認為是估計正確,沒有任何損失;
- SVR的優化目標,是|W|最小,這樣y-x曲線的斜率最小,這個function最flat,這樣據說可以增加估計的魯棒性。
- 之後的事情就很自然了,和SVM一樣:可以有soft margin,用一個小正數控制。用對偶式來解;但有一個不同,控制範圍的epsilon的值難於確定,在最小優化目標中加入一項C*\nu*\epsilon,其中epsilon是一個變數,nu是一個預先給定的正數。
四、svm設定引數
-s svm型別:SVM設定型別(預設0)
0 -- C-SVC
1 --v-SVC
2 – 一類SVM
3 -- e -SVR
4 -- v-SVR-t 核函式型別:核函式設定型別(預設2)
0 – 線性:u'v
1 – 多項式:(r*u'v + coef0)^degree
2 – RBF函式:exp(-r|u-v|^2)
3 –sigmoid:tanh(r*u'v + coef0)-d degree:核函式中的degree設定(預設3)
-g r(gama):核函式中的函式設定(預設1/ k)
-r coef0:核函式中的coef0設定(預設0)
-c cost:設定C-SVC, -SVR和-SVR的引數(預設1)
-n nu:設定SVC,一類SVM和 SVR的引數(預設0.5)
-p e:設定 -SVR 中損失函式的值(預設0.1)
-m cachesize:設定cache記憶體大小,以MB為單位(預設40)
-e :設定允許的終止判據(預設0.001)
-h shrinking:是否使用啟發式,0或1(預設1)
-wi weight:設定第幾類的引數C為weightC(C-SVC中的C)(預設1)
-v n: n-fold互動檢驗模式
五、svm模型引數
- svm_type:所選擇的svm型別,預設為c_svc
- kernel_type rbf:訓練採用的核函式型別,此處為RBF核
- gamma 0.0078125:RBF核的引數γ
- nr_class 6:類別數,此處為6分類問題
- total_sv 18:支援向量總個數
- rho 0.004423136341674322 -0.02055338568924989 0.03588086612165208 0.24771746047322893 0.00710699773513259 -0.008734834466328766 0.02297409269106355 0.24299467083662166 -0.07400614425237287 -0.0050679463881033344 0.18446534035305884 0.004123018419961004 0.22127259896446397 -0.012677989710344693 -0.2178023679167552 :判決函式的偏置項b
- label 0 9 99 999 100 101:原始檔案中的類別標識
- nr_sv 2 2 3 3 4 4:每個類的支援向量機的個數
- SV :以下為各個類的權係數及相應的支援向量
六、分類樣例
七、參考連結
八、具體程式碼
public class SvmTest3 {
public static void main(String[] args) {
String []arg ={ "trainfile/train1.txt", //存放SVM訓練模型用的資料的路徑
"trainfile/model_r.txt"}; //存放SVM通過訓練資料訓練出來的模型的路徑
String []parg={"trainfile/test2.txt", //這個是存放測試資料
"trainfile/model_r.txt", //呼叫的是訓練以後的模型
"trainfile/out_r.txt"}; //生成的結果的檔案的路徑
System.out.println("........SVM執行開始..........");
//建立一個訓練物件
SvmTrain t = new SvmTrain();
//建立一個預測或者分類的物件
SvmPredict p= new SvmPredict();
//歸一化
SvmScale svm_scale = new SvmScale();
try {
//String[] testArgs = {"-l","0", "-u","1","-s","trainfile/trainscale.txt","trainfile/train.txt"};
//svm_scale.main(testArgs);
//String[] argvScaleTest ={"-r","trainfile/trainscale.txt","trainfile/train.txt"};
//svm_scale.main(argvScaleTest);
t.main(arg); //呼叫
p.main(parg); //呼叫
} catch (IOException e) {
e.printStackTrace();
}
}
}
/**歸一化呼叫示例
* String[] testArgs = {"-l","0", "-u","1","-s","chao-test-scale","UCI-breast-cancer-tra"};
svm_scale.main(testArgs);
String[] argvScaleTest ={"-r","chao-test-scale","UCI-breast-cancer-test"};
svm_scale.main(testArgs);
svm_scale無直接生成歸一化後的檔案方法,控制檯實現命令 :
java svm_scale -s chao-test-scale train>train.scale
java svm_scale -s chao-test-scale test>test.scale
*/public class SvmTrain {
private svm_parameter param; // set by parse_command_line
private svm_problem prob; // set by read_problem
private svm_model model;
private String input_file_name; // set by parse_command_line
private String model_file_name; // set by parse_command_line
private String error_msg;
private int cross_validation;
private int nr_fold;
private static svm_print_interface svm_print_null = new svm_print_interface()
{
public void print(String s) {}
};
private static void exit_with_help()
{
System.out.print(
"Usage: svm_train [options] training_set_file [model_file]\n"
+"options:\n"
+"-s svm_type : set type of SVM (default 0)\n"
+" 0 -- C-SVC (multi-class classification)\n"
+" 1 -- nu-SVC (multi-class classification)\n"
+" 2 -- one-class SVM\n"
+" 3 -- epsilon-SVR (regression)\n"
+" 4 -- nu-SVR (regression)\n"
+"-t kernel_type : set type of kernel function (default 2)\n"
+" 0 -- linear: u'*v\n"
+" 1 -- polynomial: (gamma*u'*v + coef0)^degree\n"
+" 2 -- radial basis function: exp(-gamma*|u-v|^2)\n"
+" 3 -- sigmoid: tanh(gamma*u'*v + coef0)\n"
+" 4 -- precomputed kernel (kernel values in training_set_file)\n"
+"-d degree : set degree in kernel function (default 3)\n"
+"-g gamma : set gamma in kernel function (default 1/num_features)\n"
+"-r coef0 : set coef0 in kernel function (default 0)\n"
+"-c cost : set the parameter C of C-SVC, epsilon-SVR, and nu-SVR (default 1)\n"
+"-n nu : set the parameter nu of nu-SVC, one-class SVM, and nu-SVR (default 0.5)\n"
+"-p epsilon : set the epsilon in loss function of epsilon-SVR (default 0.1)\n"
+"-m cachesize : set cache memory size in MB (default 100)\n"
+"-e epsilon : set tolerance of termination criterion (default 0.001)\n"
+"-h shrinking : whether to use the shrinking heuristics, 0 or 1 (default 1)\n"
+"-b probability_estimates : whether to train a SVC or SVR model for probability estimates, 0 or 1 (default 0)\n"
+"-wi weight : set the parameter C of class i to weight*C, for C-SVC (default 1)\n"
+"-v n : n-fold cross validation mode\n"
+"-q : quiet mode (no outputs)\n"
);
System.exit(1);
}
private void do_cross_validation()
{
int i;
int total_correct = 0;
double total_error = 0;
double sumv = 0, sumy = 0, sumvv = 0, sumyy = 0, sumvy = 0;
double[] target = new double[prob.l];
libsvm.svm.svm_cross_validation(prob, param, nr_fold, target);
if(param.svm_type == svm_parameter.EPSILON_SVR ||
param.svm_type == svm_parameter.NU_SVR)
{
for(i=0;i<prob.l;i++)
{
double y = prob.y[i];
double v = target[i];
total_error += (v-y)*(v-y);
sumv += v;
sumy += y;
sumvv += v*v;
sumyy += y*y;
sumvy += v*y;
}
System.out.print("Cross Validation Mean squared error = "+total_error/prob.l+"\n");
System.out.print("Cross Validation Squared correlation coefficient = "+
((prob.l*sumvy-sumv*sumy)*(prob.l*sumvy-sumv*sumy))/
((prob.l*sumvv-sumv*sumv)*(prob.l*sumyy-sumy*sumy))+"\n"
);
}
else
{
for(i=0;i<prob.l;i++)
if(target[i] == prob.y[i])
++total_correct;
System.out.print("Cross Validation Accuracy = "+100.0*total_correct/prob.l+"%\n");
}
}
private void run(String argv[]) throws IOException
{
parse_command_line(argv);
read_problem();
error_msg = libsvm.svm.svm_check_parameter(prob, param);
if(error_msg != null)
{
System.err.print("ERROR: "+error_msg+"\n");
System.exit(1);
}
if(cross_validation != 0)
{
do_cross_validation();
}
else
{
model = libsvm.svm.svm_train(prob, param);
libsvm.svm.svm_save_model(model_file_name, model);
}
}
public static void main(String argv[]) throws IOException
{
SvmTrain t = new SvmTrain();
t.run(argv);
}
private static double atof(String s)
{
double d = Double.valueOf(s).doubleValue();
if (Double.isNaN(d) || Double.isInfinite(d))
{
System.err.print("NaN or Infinity in input\n");
System.exit(1);
}
return(d);
}
private static int atoi(String s)
{
return Integer.parseInt(s);
}
private void parse_command_line(String argv[])
{
int i;
svm_print_interface print_func = null; // default printing to stdout
param = new svm_parameter();
// default values
//param.svm_type = svm_parameter.C_SVC;
//param.kernel_type = svm_parameter.RBF;
//param.svm_type = svm_parameter.NU_SVR;
//param.kernel_type = svm_parameter.POLY;
param.svm_type = svm_parameter.C_SVC;
param.kernel_type = svm_parameter.POLY;
param.degree = 3;
param.gamma = 0; // 1/num_features
param.coef0 = 0;
param.nu = 0.5;
param.cache_size = 100;
param.C = 1;
param.eps = 1e-3;
param.p = 0.1;
param.shrinking = 1;
param.probability = 0;
param.nr_weight = 0;
param.weight_label = new int[0];
param.weight = new double[0];
cross_validation = 0;
// parse options
for(i=0;i<argv.length;i++)
{
if(argv[i].charAt(0) != '-') break;
if(++i>=argv.length)
exit_with_help();
switch(argv[i-1].charAt(1))
{
case 's':
param.svm_type = atoi(argv[i]);
break;
case 't':
param.kernel_type = atoi(argv[i]);
break;
case 'd':
param.degree = atoi(argv[i]);
break;
case 'g':
param.gamma = atof(argv[i]);
break;
case 'r':
param.coef0 = atof(argv[i]);
break;
case 'n':
param.nu = atof(argv[i]);
break;
case 'm':
param.cache_size = atof(argv[i]);
break;
case 'c':
param.C = atof(argv[i]);
break;
case 'e':
param.eps = atof(argv[i]);
break;
case 'p':
param.p = atof(argv[i]);
break;
case 'h':
param.shrinking = atoi(argv[i]);
break;
case 'b':
param.probability = atoi(argv[i]);
break;
case 'q':
print_func = svm_print_null;
i--;
break;
case 'v':
cross_validation = 1;
nr_fold = atoi(argv[i]);
if(nr_fold < 2)
{
System.err.print("n-fold cross validation: n must >= 2\n");
exit_with_help();
}
break;
case 'w':
++param.nr_weight;
{
int[] old = param.weight_label;
param.weight_label = new int[param.nr_weight];
System.arraycopy(old,0,param.weight_label,0,param.nr_weight-1);
}
{
double[] old = param.weight;
param.weight = new double[param.nr_weight];
System.arraycopy(old,0,param.weight,0,param.nr_weight-1);
}
param.weight_label[param.nr_weight-1] = atoi(argv[i-1].substring(2));
param.weight[param.nr_weight-1] = atof(argv[i]);
break;
default:
System.err.print("Unknown option: " + argv[i-1] + "\n");
exit_with_help();
}
}
svm.svm_set_print_string_function(print_func);
// determine filenames
if(i>=argv.length)
exit_with_help();
input_file_name = argv[i];
if(i<argv.length-1)
model_file_name = argv[i+1];
else
{
int p = argv[i].lastIndexOf('/');
++p; // whew...
model_file_name = argv[i].substring(p)+".model";
}
}
// read in a problem (in svmlight format)
private void read_problem() throws IOException
{
BufferedReader fp = new BufferedReader(new FileReader(input_file_name));
Vector<Double> vy = new Vector<Double>();
Vector<svm_node[]> vx = new Vector<svm_node[]>();
int max_index = 0;
while(true)
{
String line = fp.readLine();
if(line == null) break;
StringTokenizer st = new StringTokenizer(line," \t\n\r\f:");
vy.addElement(atof(st.nextToken()));
int m = st.countTokens()/2;
svm_node[] x = new svm_node[m];
for(int j=0;j<m;j++)
{
x[j] = new svm_node();
x[j].index = atoi(st.nextToken());
x[j].value = atof(st.nextToken());
}
if(m>0) max_index = Math.max(max_index, x[m-1].index);
vx.addElement(x);
}
prob = new svm_problem();
prob.l = vy.size();
prob.x = new svm_node[prob.l][];
for(int i=0;i<prob.l;i++)
prob.x[i] = vx.elementAt(i);
prob.y = new double[prob.l];
for(int i=0;i<prob.l;i++)
prob.y[i] = vy.elementAt(i);
if(param.gamma == 0 && max_index > 0)
param.gamma = 1.0/max_index;
if(param.kernel_type == svm_parameter.PRECOMPUTED)
for(int i=0;i<prob.l;i++)
{
if (prob.x[i][0].index != 0)
{
System.err.print("Wrong kernel matrix: first column must be 0:sample_serial_number\n");
System.exit(1);
}
if ((int)prob.x[i][0].value <= 0 || (int)prob.x[i][0].value > max_index)
{
System.err.print("Wrong input format: sample_serial_number out of range\n");
System.exit(1);
}
}
fp.close();
}
}public class SvmPredict {
private static svm_print_interface svm_print_null = new svm_print_interface()
{
public void print(String s) {}
};
private static svm_print_interface svm_print_stdout = new svm_print_interface()
{
public void print(String s)
{
System.out.print(s);
}
};
private static svm_print_interface svm_print_string = svm_print_stdout;
static void info(String s)
{
svm_print_string.print(s);
}
private static double atof(String s)
{
return Double.valueOf(s).doubleValue();
}
private static int atoi(String s)
{
return Integer.parseInt(s);
}
private static void predict(BufferedReader input, DataOutputStream output, svm_model model, int predict_probability) throws IOException
{
int correct = 0;
int total = 0;
double error = 0;
double sumv = 0, sumy = 0, sumvv = 0, sumyy = 0, sumvy = 0;
int svm_type= libsvm.svm.svm_get_svm_type(model);
int nr_class= libsvm.svm.svm_get_nr_class(model);
double[] prob_estimates=null;
if(predict_probability == 1)
{
if(svm_type == svm_parameter.EPSILON_SVR ||
svm_type == svm_parameter.NU_SVR)
{
SvmPredict.info("Prob. model for test data: target value = predicted value + z,\nz: Laplace distribution e^(-|z|/sigma)/(2sigma),sigma="+ libsvm.svm.svm_get_svr_probability(model)+"\n");
}
else
{
int[] labels=new int[nr_class];
libsvm.svm.svm_get_labels(model, labels);
prob_estimates = new double[nr_class];
output.writeBytes("labels");
for(int j=0;j<nr_class;j++)
output.writeBytes(" "+labels[j]);
output.writeBytes("\n");
}
}
while(true)
{
String line = input.readLine();
if(line == null) break;
StringTokenizer st = new StringTokenizer(line," \t\n\r\f:");
double target = atof(st.nextToken());
int m = st.countTokens()/2;
svm_node[] x = new svm_node[m];
for(int j=0;j<m;j++)
{
x[j] = new svm_node();
x[j].index = atoi(st.nextToken());
x[j].value = atof(st.nextToken());
}
double v;
if (predict_probability==1 && (svm_type==svm_parameter.C_SVC || svm_type==svm_parameter.NU_SVC))
{
v = libsvm.svm.svm_predict_probability(model, x, prob_estimates);
output.writeBytes(v+" ");
for(int j=0;j<nr_class;j++)
output.writeBytes(prob_estimates[j]+" ");
output.writeBytes("\n");
}
else
{
v = libsvm.svm.svm_predict(model, x);
output.writeBytes(v+"\n");
}
if(v == target)
++correct;
error += (v-target)*(v-target);
sumv += v;
sumy += target;
sumvv += v*v;
sumyy += target*target;
sumvy += v*target;
++total;
}
if(svm_type == svm_parameter.EPSILON_SVR ||
svm_type == svm_parameter.NU_SVR)
{
SvmPredict.info("Mean squared error = "+error/total+" (regression)\n");
SvmPredict.info("Squared correlation coefficient = "+
((total*sumvy-sumv*sumy)*(total*sumvy-sumv*sumy))/
((total*sumvv-sumv*sumv)*(total*sumyy-sumy*sumy))+
" (regression)\n");
}
else
SvmPredict.info("Accuracy = "+(double)correct/total*100+
"% ("+correct+"/"+total+") (classification)\n");
}
private static void exit_with_help()
{
System.err.print("usage: svm_predict [options] test_file model_file output_file\n"
+"options:\n"
+"-b probability_estimates: whether to predict probability estimates, 0 or 1 (default 0); one-class SVM not supported yet\n"
+"-q : quiet mode (no outputs)\n");
System.exit(1);
}
public static void main(String argv[]) throws IOException
{
int i, predict_probability=0;
svm_print_string = svm_print_stdout;
// parse options
for(i=0;i<argv.length;i++)
{
if(argv[i].charAt(0) != '-') break;
++i;
switch(argv[i-1].charAt(1))
{
case 'b':
predict_probability = atoi(argv[i]);
break;
case 'q':
svm_print_string = svm_print_null;
i--;
break;
default:
System.err.print("Unknown option: " + argv[i-1] + "\n");
exit_with_help();
}
}
if(i>=argv.length-2)
exit_with_help();
try
{
BufferedReader input = new BufferedReader(new FileReader(argv[i]));
DataOutputStream output = new DataOutputStream(new BufferedOutputStream(new FileOutputStream(argv[i+2])));
svm_model model = libsvm.svm.svm_load_model(argv[i + 1]);
if (model == null)
{
System.err.print("can't open model file "+argv[i+1]+"\n");
System.exit(1);
}
if(predict_probability == 1)
{
if(libsvm.svm.svm_check_probability_model(model)==0)
{
System.err.print("Model does not support probabiliy estimates\n");
System.exit(1);
}
}
else
{
if(svm.svm_check_probability_model(model)!=0)
{
SvmPredict.info("Model supports probability estimates, but disabled in prediction.\n");
}
}
predict(input,output,model,predict_probability);
input.close();
output.close();
}
catch(FileNotFoundException e)
{
exit_with_help();
}
catch(ArrayIndexOutOfBoundsException e)
{
exit_with_help();
}
}
}public class SvmScale {
private String line = null;
private double lower = -1.0;
private double upper = 1.0;
private double y_lower;
private double y_upper;
private boolean y_scaling = false;
private double[] feature_max;
private double[] feature_min;
private double y_max = -Double.MAX_VALUE;
private double y_min = Double.MAX_VALUE;
private int max_index;
private long num_nonzeros = 0;
private long new_num_nonzeros = 0;
private static void exit_with_help()
{
System.out.print(
"Usage: svm-scale [options] data_filename\n"
+"options:\n"
+"-l lower : x scaling lower limit (default -1)\n"
+"-u upper : x scaling upper limit (default +1)\n"
+"-y y_lower y_upper : y scaling limits (default: no y scaling)\n"
+"-s save_filename : save scaling parameters to save_filename\n"
+"-r restore_filename : restore scaling parameters from restore_filename\n"
);
System.exit(1);
}
private BufferedReader rewind(BufferedReader fp, String filename) throws IOException
{
fp.close();
return new BufferedReader(new FileReader(filename));
}
private void output_target(double value)
{
if(y_scaling)
{
if(value == y_min)
value = y_lower;
else if(value == y_max)
value = y_upper;
else
value = y_lower + (y_upper-y_lower) *
(value-y_min) / (y_max-y_min);
}
System.out.print(value + " ");
}
private void output(int index, double value)
{
/* skip single-valued attribute */
if(feature_max[index] == feature_min[index])
return;
if(value == feature_min[index])
value = lower;
else if(value == feature_max[index])
value = upper;
else
value = lower + (upper-lower) *
(value-feature_min[index])/
(feature_max[index]-feature_min[index]);
if(value != 0)
{
System.out.print(index + ":" + value + " ");
new_num_nonzeros++;
}
}
private String readline(BufferedReader fp) throws IOException
{
line = fp.readLine();
return line;
}
private void run(String []argv) throws IOException
{
int i,index;
BufferedReader fp = null, fp_restore = null;
String save_filename = null;
String restore_filename = null;
String data_filename = null;
for(i=0;i<argv.length;i++)
{
if (argv[i].charAt(0) != '-') break;
++i;
switch(argv[i-1].charAt(1))
{
case 'l': lower = Double.parseDouble(argv[i]); break;
case 'u': upper = Double.parseDouble(argv[i]); break;
case 'y':
y_lower = Double.parseDouble(argv[i]);
++i;
y_upper = Double.parseDouble(argv[i]);
y_scaling = true;
break;
case 's': save_filename = argv[i]; break;
case 'r': restore_filename = argv[i]; break;
default:
System.err.println("unknown option");
exit_with_help();
}
}
if(!(upper > lower) || (y_scaling && !(y_upper > y_lower)))
{
System.err.println("inconsistent lower/upper specification");
System.exit(1);
}
if(restore_filename != null && save_filename != null)
{
System.err.println("cannot use -r and -s simultaneously");
System.exit(1);
}
if(argv.length != i+1) // modified by yehui
// if(argv.length != i)
exit_with_help();
data_filename = argv[i];// modified by yehui
//data_filename = argv[i-1];
try {
fp = new BufferedReader(new FileReader(data_filename));
} catch (Exception e) {
System.err.println("can't open file " + data_filename);
System.exit(1);
}
/* assumption: min index of attributes is 1 */
/* pass 1: find out max index of attributes */
max_index = 0;
if(restore_filename != null)
{
int idx, c;
try {
fp_restore = new BufferedReader(new FileReader(restore_filename));
}
catch (Exception e) {
System.err.println("can't open file " + restore_filename);
System.exit(1);
}
if((c = fp_restore.read()) == 'y')
{
fp_restore.readLine();
fp_restore.readLine();
fp_restore.readLine();
}
fp_restore.readLine();
fp_restore.readLine();
String restore_line = null;
while((restore_line = fp_restore.readLine())!=null)
{
StringTokenizer st2 = new StringTokenizer(restore_line);
idx = Integer.parseInt(st2.nextToken());
max_index = Math.max(max_index, idx);
}
fp_restore = rewind(fp_restore, restore_filename);
}
while (readline(fp) != null)
{
StringTokenizer st = new StringTokenizer(line," \t\n\r\f:");
st.nextToken();
while(st.hasMoreTokens())
{
try {
index = Integer.parseInt(st.nextToken());
max_index = Math.max(max_index, index);
st.nextToken();
num_nonzeros++;
} catch (NumberFormatException e){
System.out.println(e);
}
}
}
try {
feature_max = new double[(max_index+1)];
feature_min = new double[(max_index+1)];
} catch(OutOfMemoryError e) {
System.err.println("can't allocate enough memory");
System.exit(1);
}
for(i=0;i<=max_index;i++)
{
feature_max[i] = -Double.MAX_VALUE;
feature_min[i] = Double.MAX_VALUE;
}
fp = rewind(fp, data_filename);
/* pass 2: find out min/max value */
while(readline(fp) != null)
{
int next_index = 1;
double target;
double value;
StringTokenizer st = new StringTokenizer(line," \t\n\r\f:");
target = Double.parseDouble(st.nextToken());
y_max = Math.max(y_max, target);
y_min = Math.min(y_min, target);
while (st.hasMoreTokens())
{
index = Integer.parseInt(st.nextToken());
value = Double.parseDouble(st.nextToken());
for (i = next_index; i<index; i++)
{
feature_max[i] = Math.max(feature_max[i], 0);
feature_min[i] = Math.min(feature_min[i], 0);
}
feature_max[index] = Math.max(feature_max[index], value);
feature_min[index] = Math.min(feature_min[index], value);
next_index = index + 1;
}
for(i=next_index;i<=max_index;i++)
{
feature_max[i] = Math.max(feature_max[i], 0);
feature_min[i] = Math.min(feature_min[i], 0);
}
}
fp = rewind(fp, data_filename);
/* pass 2.5: save/restore feature_min/feature_max */
if(restore_filename != null)
{
// fp_restore rewinded in finding max_index
int idx, c;
double fmin, fmax;
fp_restore.mark(2); // for reset
if((c = fp_restore.read()) == 'y')
{
fp_restore.readLine(); // pass the '\n' after 'y'
StringTokenizer st = new StringTokenizer(fp_restore.readLine());
y_lower = Double.parseDouble(st.nextToken());
y_upper = Double.parseDouble(st.nextToken());
st = new StringTokenizer(fp_restore.readLine());
y_min = Double.parseDouble(st.nextToken());
y_max = Double.parseDouble(st.nextToken());
y_scaling = true;
}
else
fp_restore.reset();
if(fp_restore.read() == 'x') {
fp_restore.readLine(); // pass the '\n' after 'x'
StringTokenizer st = new StringTokenizer(fp_restore.readLine());
lower = Double.parseDouble(st.nextToken());
upper = Double.parseDouble(st.nextToken());
String restore_line = null;
while((restore_line = fp_restore.readLine())!=null)
{
StringTokenizer st2 = new StringTokenizer(restore_line);
idx = Integer.parseInt(st2.nextToken());
fmin = Double.parseDouble(st2.nextToken());
fmax = Double.parseDouble(st2.nextToken());
if (idx <= max_index)
{
feature_min[idx] = fmin;
feature_max[idx] = fmax;
}
}
}
fp_restore.close();
}
if(save_filename != null)
{
Formatter formatter = new Formatter(new StringBuilder());
BufferedWriter fp_save = null;
try {
fp_save = new BufferedWriter(new FileWriter(save_filename));
} catch(IOException e) {
System.err.println("can't open file " + save_filename);
System.exit(1);
}
if(y_scaling)
{
formatter.format("y\n");
formatter.format("%.16g %.16g\n", y_lower, y_upper);
formatter.format("%.16g %.16g\n", y_min, y_max);
}
formatter.format("x\n");
formatter.format("%.16g %.16g\n", lower, upper);
for(i=1;i<=max_index;i++)
{
if(feature_min[i] != feature_max[i])
formatter.format("%d %.16g %.16g\n", i, feature_min[i], feature_max[i]);
}
fp_save.write(formatter.toString());
fp_save.close();
}
/* pass 3: scale */
while(readline(fp) != null)
{
int next_index = 1;
double target;
double value;
StringTokenizer st = new StringTokenizer(line," \t\n\r\f:");
target = Double.parseDouble(st.nextToken());
output_target(target);
while(st.hasMoreElements())
{
index = Integer.parseInt(st.nextToken());
value = Double.parseDouble(st.nextToken());
for (i = next_index; i<index; i++)
output(i, 0);
output(index, value);
next_index = index + 1;
}
for(i=next_index;i<= max_index;i++)
output(i, 0);
System.out.print("\n");
}
if (new_num_nonzeros > num_nonzeros)
System.err.print(
"WARNING: original #nonzeros " + num_nonzeros+"\n"
+" new #nonzeros " + new_num_nonzeros+"\n"
+"Use -l 0 if many original feature values are zeros\n");
fp.close();
}
public static void main(String argv[]) throws IOException
{
SvmScale s = new SvmScale();
s.run(argv);
}
}public class SvmToy extends Applet {
static final String DEFAULT_PARAM="-t 2 -c 100";
int XLEN;
int YLEN;
// off-screen buffer
Image buffer;
Graphics buffer_gc;
// pre-allocated colors
final static Color colors[] =
{
new Color(0,0,0),
new Color(0,120,120),
new Color(120,120,
相關推薦
機器學習-多分類問題
一、背景
為何選擇svm來嘗試解決多分類問題,以下為決策樹和svm用於多分類的表現上的差異:
二、SVM支援向量機(Support vector machine)
設計k個SVM兩類分類器;
設計兩兩k(k-1)/2個SVM兩類分類器。
線上
Sklearn,xgboost機器學習多分類實驗
一. 背景
多分類是一個機器學習的常見任務,本文將基於復旦大學中文文字分類語料,使用sklearn和xgboost來進行多分類實驗。
預裝軟體包:
1. Jieba分詞:
環境: linux fedora 23
2. Sklearn:
3.xgboost:
安裝好
吳恩達-機器學習(3)-分類、邏輯迴歸、多分類、過擬合
文章目錄
Classification and Representation
Classification
Hypothesis Representation
Decision Boundary
機器學習之分類問題實戰(基於UCI Bank Marketing Dataset)
表示 般的 機構 文件 cnblogs opened csv文件 mas htm
導讀:
分類問題是機器學習應用中的常見問題,而二分類問題是其中的典型,例如垃圾郵件的識別。本文基於UCI機器學習數據庫中的銀行營銷數據集,從對數據集進行探索,數據預處理和特征工程,到學習
機器學習-KNN分類器
pos show sha key borde 不同 簡單的 測試 solid 1. K-近鄰(k-Nearest Neighbors,KNN)的原理
通過測量不同特征值之間的距離來衡量相似度的方法進行分類。
2. KNN算法過程
訓練樣本集:樣本集中每個特征值都
機器學習二——分類算法--決策樹DecisionTree
其中 .cn 比較 輸出 選擇 結構 沒有 ati 流程圖
機器學習算法評估標準:準確率,速度,強壯性(噪音影響較小),可規模性,可解釋性。
1、決策樹 Decision Tree:決策樹是一個類似於流程圖的樹結構,其中每個內部節點表示在一個屬性上的測試,每一個分支代表
機器學習三--分類--鄰近取樣(Nearest Neighbor)
post 個數 均衡 urn learning clas 根據 () end 最鄰近規則分類 K-Nearest Neighbor
步驟:
1、為了判斷未知實例的類別,以所有已知類別的實例作為參考。
2、選擇參數K。
3、計算未知實例與所有已知實例的距離。
機器學習(四) 分類算法--K近鄰算法 KNN
class 給定 sort sta shape counter 3.5 解釋 sqrt 一、K近鄰算法基礎
KNN------- K近鄰算法--------K-Nearest Neighbors
思想極度簡單
應用數學知識少 (近乎為零)
效果好(缺點?)
可以解
機器學習的分類與主要演算法對比
重要引用:Andrew Ng Courera Machine Learning;從機器學習談起;關於機器學習的討論;機器學習常見演算法分類彙總;LeNet Homepage;pluskid svm
首先讓我們瞻仰一下當今機器學習領域的執牛耳者:
這幅圖上的三人是當今機器學習界的
吳恩達機器學習-多變數線性迴歸 吳恩達機器學習 - 多變數線性迴歸
原
吳恩達機器學習 - 多變數線性迴歸
2018年06月18日 17:50:26
離殤灬孤狼
閱讀數:84
收起
機器學習之分類和迴歸區別闡述
很多人分不清楚分類和迴歸,我來講一下,我們經常會碰到這樣的問題:
1、如何將信用卡申請人分為低、中、高風險群?
2、如何預測哪些顧客在未來半年內會取消該公司服務,哪些電話使用者會申請增值服務?
3、如何預測具有某些特徵的顧客是否會購買一臺新的計算機?
4、如何預測病人應當接受三種
【機器學習】分類決策樹與迴歸決策樹案例
一、回顧
什麼是決策樹,資訊熵
構建決策樹的過程
ID3、C4.5和CRAT演算法
上面三篇,主要介紹了相關的理論知識,其中構建決策樹的過程可以很好地幫助我們理解決策樹的分裂屬性的選擇。
本篇所有原始碼:Github
二
機器學習之分類決策樹DecisionTreeClassifier
機器學習之分類決策樹DecisionTreeClassifier
# -*- coding: utf-8 -*-
"""
Created on Fri Nov 23 21:06:54 2018
@author: muli
"""
import numpy as np
機器學習中分類與迴歸的解決與區別
機器學習可以解決很多問題,其中最為重要的兩個是 迴歸與分類。 這兩個問題怎麼解決, 它們之間又有什麼區別呢? 以下舉幾個簡單的例子,以給大家一個概念
1. 線性迴歸
迴歸分析常用於分析兩個變數X和Y 之間的關係。 比如 X=房子大小 和 Y=房價 之間的關係, X=(公園人流量,公園門票票價
【深度學習-機器學習】分類度量指標 : 正確率、召回率、靈敏度、特異度,ROC曲線、AUC等
在分類任務中,人們總是喜歡基於錯誤率來衡量分類器任務的成功程度。錯誤率指的是在所有測試樣例中錯分的樣例比例。實際上,這樣的度量錯誤掩蓋了樣例如何被分錯的事實。在機器學習中,有一個普遍適用的稱為混淆矩陣(confusion matrix)的工具,它可以幫助人們更好地瞭解
機器學習SVC分類預測三個月後的股價
思路:通過學習近兩年的每個季度報的基本面財務資料,建立模型,買入並持有預測三個月後會漲5%以上的股票,直到下一批季度報
資料採集:用到了大約10018行資料(已去除缺失值,不採用填充),其中採用了兩個技術指標(趨勢指標CYES,CYEL)
circulating_market_ca
機器學習中分類與迴歸問題的區別與聯絡
分類和迴歸問題之間存在重要差異。
從根本上說,分類是關於預測標籤,而回歸是關於預測數量。
我經常看到諸如以下問題:
如何計算迴歸問題的準確性?
像這樣的問題是沒有真正理解分類和迴歸之間的差異以及試圖衡量的準確度的症狀。
在本教程中,您將發現分類和迴歸之間的差異。
機器學習3-分類及模型驗證
目錄
機器分類
人工分類
二元分類
多元分類
樸素貝葉斯分類
訓練相關
劃分訓練集和測試集
交叉驗證
混淆矩陣
分類報告
基於決策樹的分類
基於投
機器學習之分類問題的效能度量
機器學習之分類問題的效能度量
# -*- coding: utf-8 -*-
"""
Created on Mon Dec 10 10:54:09 2018
@author: muli
"""
from sklearn.metrics import accuracy
機器學習---線性分類器三種最優準則
線性分類器三種最優準則: Fisher 準則 :根據兩類樣本一般類內密集,類間分離的特點,尋找線性分類器最佳的法線向量方向,使兩類樣本在該方向上的投影滿足類內儘可能密集,類間儘可能分開。這種度量通過類內