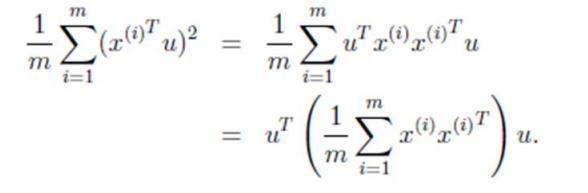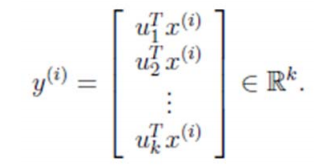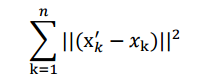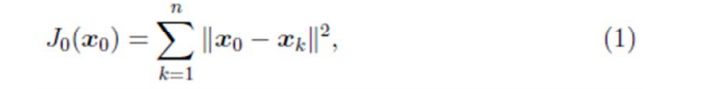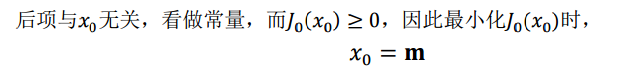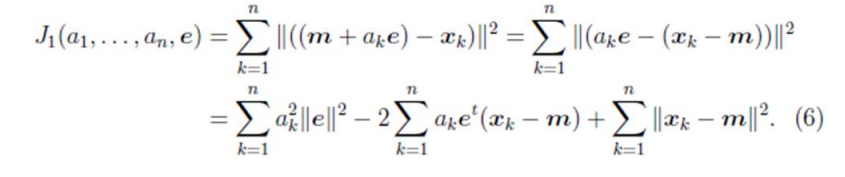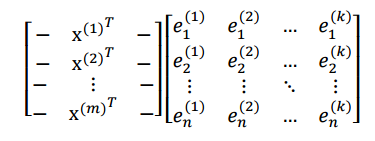主成分分析(PCA)-理論基礎
要解釋為什麼協方差矩陣的特徵向量可以將原始特徵對映到 k 維理想特徵,我看到的有三個理論:分別是最大方差理論、最小錯誤理論和座標軸相關度理論。這裡簡單探討前兩種,最後一種在討論PCA 意義時簡單概述。
最大方差理論
在訊號處理中認為訊號具有較大的方差,噪聲有較小的方差,信噪比就是訊號與噪聲的方差比,越大越好。如前面的圖,樣本在橫軸上的投影方差較大,在縱軸上的投影方差較小,那麼認為縱軸上的投影是由噪聲引起的。
因此我們認為,最好的 k 維特徵是將 n 維樣本點轉換為 k 維後,每一維上的樣本方差都很大。
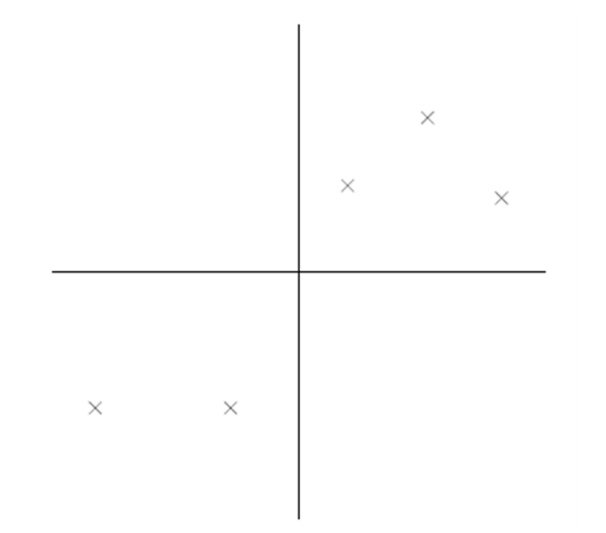
比如下圖有 5 個樣本點:(已經做過預處理,均值為 0,特徵方差歸一)
下面將樣本投影到某一維上,這裡用一條過原點的直線表示(之前預處理的過程實質是將原點移到樣本點的中心點)。
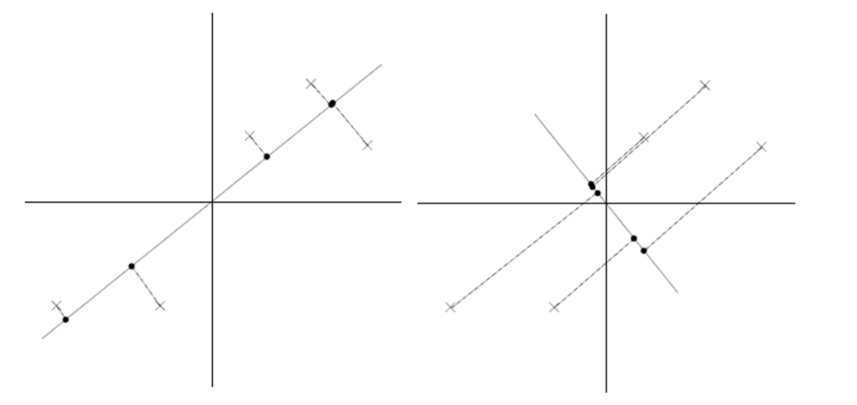
假設我們選擇兩條不同的直線做投影,那麼左右兩條中哪個好呢?根據我們之前的方差最大化理論,左邊的好,因為投影后的樣本點之間方差最大。
這裡先解釋一下投影的概念:
紅色點表示樣例
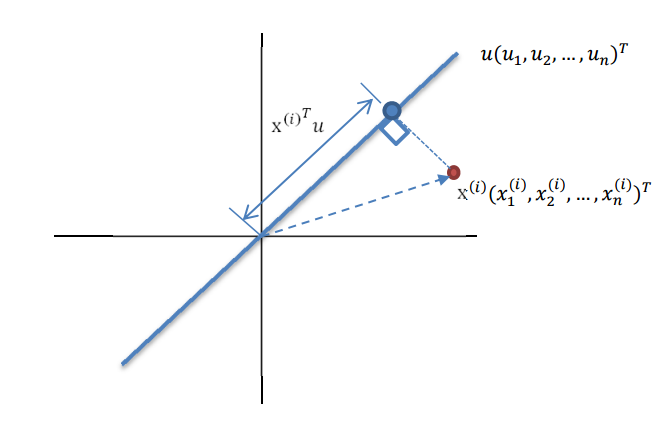
回到上面左右圖中的左圖,我們要求的是最佳的 u,使得投影后的樣本點方差最大。
由於投影后均值為 0,因此方差為:
中間那部分很熟悉啊,不就是樣本特徵的協方差矩陣麼(
We got it!λ就是Σ的特徵值,u 是特徵向量。最佳的投影直線是特徵值λ最大時對應的特徵向量,其次是λ第二大對應的特徵向量,依次類推。
因此,我們只需要對協方差矩陣進行特徵值分解,得到的前 k 大特徵值對應的特徵向量就是最佳的 k 維新特徵,而且這 k 維新特徵是正交的。得到前 k 個 u 以後,樣例
通過選取最大的 k 個 u,使得方差較小的特徵(如噪聲)被丟棄。
這是其中一種對 PCA 的解釋,第二種是錯誤最小化。
最小平方誤差理論
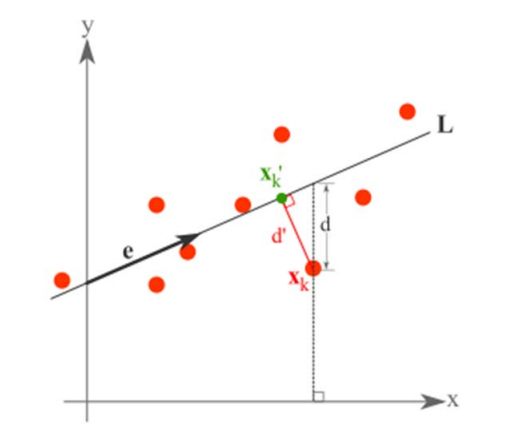
假設有這樣的二維樣本點(紅色點),回顧我們前面探討的是求一條直線,使得樣本點投影到直線上的點的方差最大。本質是求直線,那麼度量直線求的好不好,不僅僅只有方差最大化的方法。再回想我們最開始學習的線性迴歸等,目的也是求一個線性函式使得直線能夠最佳擬合樣本點,那麼我們能不能認為最佳的直線就是迴歸後的直線呢?迴歸時我們的最小二乘法度量的是樣本點到直線的座標軸距離。比如這個問題中,特徵是 x,類標籤是 y。迴歸時最小二乘法度量的是距離 d。如果使用迴歸方法來度量最佳直線,那麼就是直接在原始樣本上做迴歸了,跟特徵選擇就沒什麼關係了。
因此,我們打算選用另外一種評價直線好壞的方法,使用點到直線的距離 d’來度量。
現在有 n 個樣本點(
這個公式稱作最小平方誤差(Least Squared Error)。
而確定一條直線,一般只需要確定一個點以及方向即可。
第一步確定點:
假設要在空間中找一點
最小。其中
那麼平方錯誤可以寫作:
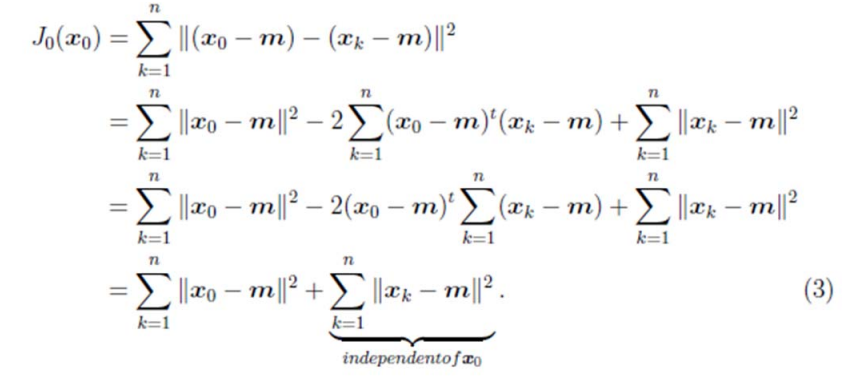
第一行就類似座標系移動到以m為中心。
因此我們選取
第二步確定方向:
我們首先固定 e,將其看做是常量,||e||=1,然後對
這個結果意思是說,如果知道了 e,那麼將

兩邊除以 n‐1 就變成了,對協方差矩陣求特徵值向量了。
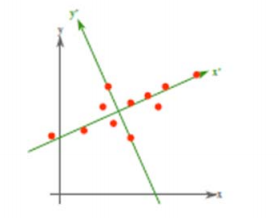
從不同的思路出發,最後得到同一個結果,對協方差矩陣求特徵向量,求得後特徵向量上就成為了新的座標,如下圖:
這時候點都聚集在新的座標軸周圍,因為我們使用的最小平方誤差的意義就在此。
PCA 理論意義
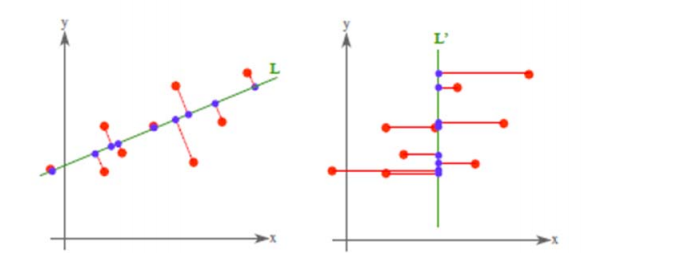
PCA 將 n 個特徵降維到 k 個,可以用來進行資料壓縮,如果 100 維的向量最後可以用10 維來表示,那麼壓縮率為 90%。同樣影象處理領域的 KL 變換使用 PCA 做影象壓縮。但PCA 要保證降維後,還要保證資料的特性損失最小。再看回顧一下 PCA 的效果。經過 PCA處理後,二維資料投影到一維上可以有以下幾種情況:
我們認為左圖好,一方面是投影后方差最大,一方面是點到直線的距離平方和最小,而且直線過樣本點的中心點。為什麼右邊的投影效果比較差?直覺是因為座標軸之間相關,以至於去掉一個座標軸,就會使得座標點無法被單獨一個座標軸確定。
PCA 得到的 k 個座標軸實際上是 k 個特徵向量,由於協方差矩陣對稱,因此 k 個特徵向量正交。看下面的計算過程。
我們最後得到的投影結果是AE,E 是 k 個特徵向量組成的矩陣,展開如下:
得到的新的樣例矩陣就是 m 個樣例到 k 個特徵向量的投影,也是這 k 個特徵向量的線性組合。。e 之間是正交的。從矩陣乘法中可以看出,PCA 所做的變換是將原始樣本點(n 維),投影到 k 個正交的座標系中去,丟棄其他維度的資訊。
舉個例子,假設宇宙是 n 維的(霍金說是 13 維的),我們得到銀河系中每個星星的座標(相對於銀河系中心的 n 維向量),然而我們想用二維座標去逼近這些樣本點,假設算出來的協方差矩陣的特徵向量分別是圖中的水平和豎直方向,那麼我們建議以銀河系中心為原點的 x 和 y 座標軸,所有的星星都投影到 x和 y 上,得到下面的圖片。然而我們丟棄了每個星星離我們的遠近距離等資訊。
總結與討論
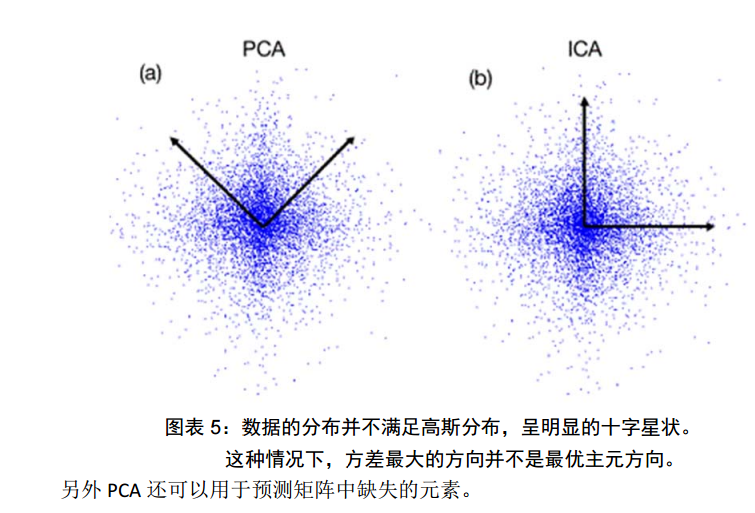
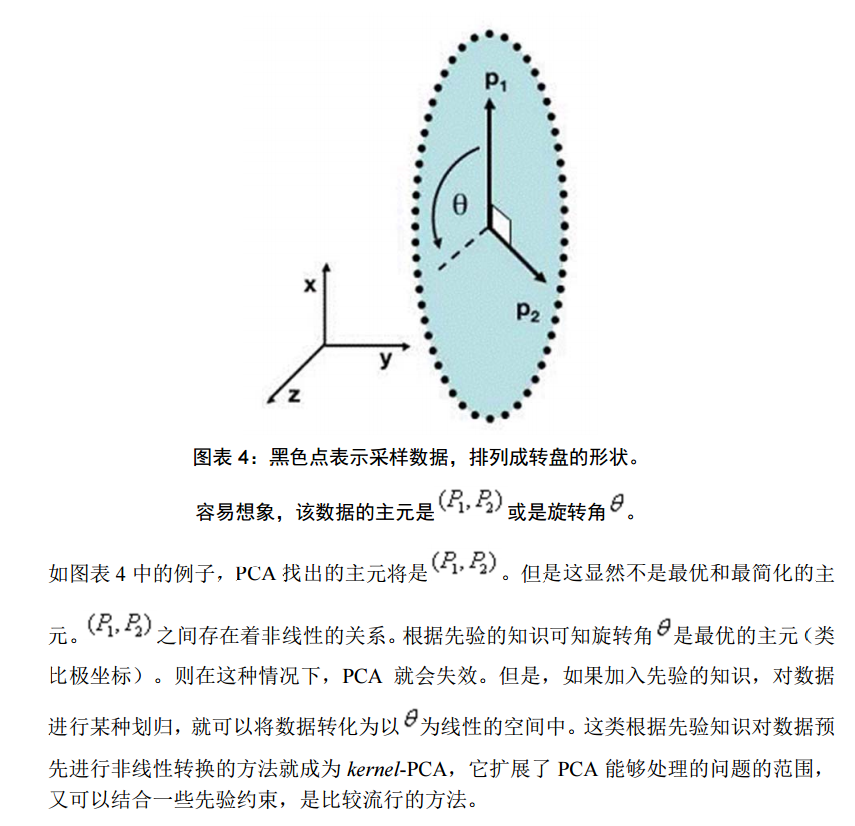
但是,這一點同時也可以看作是缺點。如果使用者對觀測物件有一定的先驗知識,掌握了資料的一些特徵,卻無法通過引數化等方法對處理過程進行干預,可能會得不到預期的效果,效率也不高。

其他參考文獻
下面的練習來自UFLDL
pca2d
close all
%%================================================================
%% Step 0: Load data
% We have provided the code to load data from pcaData.txt into x.
% x is a 2 * 45 matrix, where the kth column x(:,k) corresponds to
% the kth data point.Here we provide the code to load natural image data into x.
% You do not need to change the code below.
x = load('pcaData.txt','-ascii');
figure(1);
scatter(x(1, :), x(2, :));
title('Raw data');
%%================================================================
%% Step 1a: Implement PCA to obtain U
% Implement PCA to obtain the rotation matrix U, which is the eigenbasis
% sigma.
% -------------------- YOUR CODE HERE --------------------
u = zeros(size(x, 1)); % You need to compute this
avg = mean(x,2);
x = x - repmat(avg,1,size(x,2));
sigma = x * x' /size(x,2);
[U,S,V] = svd(sigma);
u = U;
% --------------------------------------------------------
hold on
plot([0 u(1,1)], [0 u(2,1)]);
plot([0 u(1,2)], [0 u(2,2)]);
scatter(x(1, :), x(2, :));
hold off
%%================================================================
%% Step 1b: Compute xRot, the projection on to the eigenbasis
% Now, compute xRot by projecting the data on to the basis defined
% by U. Visualize the points by performing a scatter plot.
% -------------------- YOUR CODE HERE --------------------
xRot = zeros(size(x)); % You need to compute this
xRot = u'*x;
% --------------------------------------------------------
% Visualise the covariance matrix. You should see a line across the
% diagonal against a blue background.
figure(2);
scatter(xRot(1, :), xRot(2, :));
title('xRot');
%%================================================================
%% Step 2: Reduce the number of dimensions from 2 to 1.
% Compute xRot again (this time projecting to 1 dimension).
% Then, compute xHat by projecting the xRot back onto the original axes
% to see the effect of dimension reduction
% -------------------- YOUR CODE HERE --------------------
k = 1; % Use k = 1 and project the data onto the first eigenbasis
xHat = zeros(size(x)); % You need to compute this
x_pca = u(:,1)' * x;
xHat = U*[x_pca;zeros(1,size(x,2))];
% --------------------------------------------------------
figure(3);
scatter(xHat(1, :), xHat(2, :));
title('xHat');
%%================================================================
%% Step 3: PCA Whitening
% Complute xPCAWhite and plot the results.
epsilon = 1e-5;
% -------------------- YOUR CODE HERE --------------------
xPCAWhite = zeros(size(x)); % You need to compute this
xPCAWhite = diag( 1./sqrt(diag(S)+epsilon) ) * xRot;
% --------------------------------------------------------
figure(4);
scatter(xPCAWhite(1, :), xPCAWhite(2, :));
title('xPCAWhite');
%%================================================================
%% Step 3: ZCA Whitening
% Complute xZCAWhite and plot the results.
% -------------------- YOUR CODE HERE --------------------
xZCAWhite = zeros(size(x)); % You need to compute this
xZCAWhite = U * xPCAWhite;
% --------------------------------------------------------
figure(5);
scatter(xZCAWhite(1, :), xZCAWhite(2, :));
title('xZCAWhite');
%% Congratulations! When you have reached this point, you are done!
% You can now move onto the next PCA exercise. :)pca_gen
%%================================================================
%% Step 0a: Load data
% Here we provide the code to load natural image data into x.
% x will be a 144 * 10000 matrix, where the kth column x(:, k) corresponds to
% the raw image data from the kth 12x12 image patch sampled.
% You do not need to change the code below.
x = sampleIMAGESRAW();
figure('name','Raw images');
randsel = randi(size(x,2),200,1); % A random selection of samples for visualization
display_network(x(:,randsel));%這裡只顯示了部分子樣本
%%================================================================
%% Step 0b: Zero-mean the data (by row)
% You can make use of the mean and repmat/bsxfun functions.
% -------------------- YOUR CODE HERE --------------------
[n,m] = size(x) %n是屬性,m是樣本數
avg_features = mean(x,2) %求得每一維特徵的均值 2代表按行求均值
x = x - repmat(avg_features, 1, size(x,2));
display_network(x(:,randsel));%做0均值前後的對比
%%================================================================
%% Step 1a: Implement PCA to obtain xRot
% Implement PCA to obtain xRot, the matrix in which the data is expressed
% with respect to the eigenbasis of sigma, which is the matrix U.
% -------------------- YOUR CODE HERE --------------------
xRot = zeros(size(x)); % You need to compute this
sigma = x*x'/size(x,2);
[U,S,V] = svd(sigma)%計算特徵向量 U特徵向量構成的矩陣,特徵向量數與屬性數相同,第一個特徵向量與某個樣本相乘解a1*x1得xRot中樣本x1的第一個屬性,S特徵值
xRot = U' * x;
%%================================================================
%% Step 1b: Check your implementation of PCA
% The covariance matrix for the data expressed with respect to the basis U
% should be a diagonal matrix with non-zero entries only along the main
% diagonal. We will verify this here.
% Write code to compute the covariance matrix, covar.
% When visualised as an image, you should see a straight line across the
% diagonal (non-zero entries) against a blue background (zero entries).
% -------------------- YOUR CODE HERE --------------------
covar = zeros(size(x, 1)); % You need to compute this
covar = xRot*xRot';%協方差是屬性間的協方差,不是每個樣本的協方差
% Visualise the covariance matrix. You should see a line across the
% diagonal against a blue background.
figure('name','Visualisation of covariance matrix');
imagesc(covar);
%%================================================================
%% Step 2: Find k, the number of components to retain
% Write code to determine k, the number of components to retain in order
% to retain at least 99% of the variance.
% -------------------- YOUR CODE HERE --------------------
k = 0; % Set k accordingly
var_sum = sum(diag(covar));%covar主對角線上就是特徵值
curr_var_sum = 0;
for i = 1:length(covar)
curr_var_sum = curr_var_sum + covar(i,i);
if curr_var_sum / var_sum >= 0.99
k=i;
break;
end
end
%%================================================================
%% Step 3: Implement PCA with dimension reduction
% Now that you have found k, you can reduce the dimension of the data by
% discarding the remaining dimensions. In this way, you can represent the
% data in k dimensions instead of the original 144, which will save you
% computational time when running learning algorithms on the reduced
% representation.
%
% Following the dimension reduction, invert the PCA transformation to produce
% the matrix xHat, the dimension-reduced data with respect to the original basis.
% Visualise the data and compare it to the raw data. You will observe that
% there is little loss due to throwing away the principal components that
% correspond to dimensions with low variation.
% -------------------- YOUR CODE HERE --------------------
xHat = zeros(size(x)); % You need to compute this
xTitle = U(:,1:k)' * x; %這個是pca降維後的結果
xHat = U*[xTitle;zeros(n-k,m)];%由pca後的資料恢復原圖
% Visualise the data, and compare it to the raw data
% You should observe that the raw and processed data are of comparable quality.
% For comparison, you may wish to generate a PCA reduced image which
% retains only 90% of the variance.
figure('name',['PCA processed images ',sprintf('(%d / %d dimensions)', k, size(x, 1)),'']);
display_network(xHat(:,randsel));
figure('name','Raw images');
display_network(x(:,randsel));
%%================================================================
%% Step 4a: Implement PCA with whitening and regularisation
% Implement PCA with whitening and regularisation to produce the matrix
% xPCAWhite.
epsilon = 0.1;
xPCAWhite = zeros(size(x));
% -------------------- YOUR CODE HERE --------------------
xPCAWhite = diag(1./sqrt(diag(S) + epsilon)) * xRot;
%%================================================================
%% Step 4b: Check your implementation of PCA whitening
% Check your implementation of PCA whitening with and without regularisation.
% PCA whitening without regularisation results a covariance matrix
% that is equal to the identity matrix. PCA whitening with regularisation
% results in a covariance matrix with diagonal entries starting close to
% 1 and gradually becoming smaller. We will verify these properties here.
% Write code to compute the covariance matrix, covar.
%
% Without regularisation (set epsilon to 0 or close to 0),
% when visualised as an image, you should see a red line across the
% diagonal (one entries) against a blue background (zero entries).
% With regularisation, you should see a red line that slowly turns
% blue across the diagonal, corresponding to the one entries slowly
% becoming smaller.
% -------------------- YOUR CODE HERE --------------------
covar = xPCAWhite * xPCAWhite';
% Visualise the covariance matrix. You should see a red line across the
% diagonal against a blue background.
figure('name','Visualisation of covariance matrix');
imagesc(covar);
%%================================================================
%% Step 5: Implement ZCA whitening
% Now implement ZCA whitening to produce the matrix xZCAWhite.
% Visualise the data and compare it to the raw data. You should observe
% that whitening results in, among other things, enhanced edges.
xZCAWhite = zeros(size(x));%ZCA白化一般都作用於所有維度,資料不降維
xZCAWhite = U * xPCAWhite; %ZCA其實就是做了PCA白化後再復原原圖---增強邊緣
% -------------------- YOUR CODE HERE --------------------
% Visualise the data, and compare it to the raw data.
% You should observe that the whitened images have enhanced edges.
figure('name','ZCA whitened images');
display_network(xZCAWhite(:,randsel));
figure('name','Raw images');
display_network(x(:,randsel));
使用pca的基本步驟是
輸入x—對x做零均值—計算x的協方差矩陣—計算特徵值特徵向量—-白化(可選,它可消除樣本間的相關性)—-選取前k個特徵向量