【ADNI】資料預處理(3)CNNs
阿新 • • 發佈:2018-11-04
ADNI Series
1、【ADNI】資料預處理(1)SPM,CAT12
2、【ADNI】資料預處理(2)獲取 subject slices
3、【ADNI】資料預處理(3)CNNs
4、【ADNI】資料預處理(4)Get top k slices according to CNNs
5、【ADNI】資料預處理(5)Get top k slices (pMCI_sMCI) according to CNNs
6、【ADNI】資料預處理(6)ADNI_slice_dataloader ||| show image
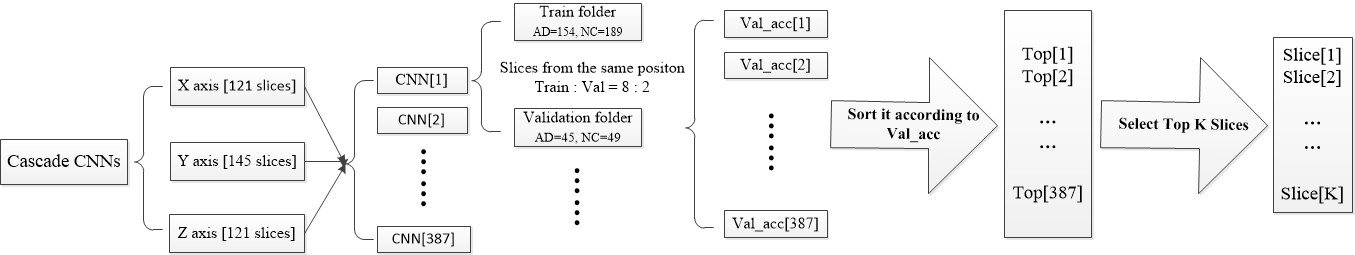
Idea:
已有資料:AD_NC_ALL_SLICE
AD_NC_ALL_SLICE:分別對ADNI下載而來的nii資料(121x145x121)沿x,y,z軸方向進行切片,得到121+145+121=387張切片圖,分別以AD/NC(199/230)為類別儲存在如下目錄:
基於上述資料形式,進一步處理:
1)目的:分別對每個切片位置組成的資料進行分類,篩選出具有區分能力的切片位置;
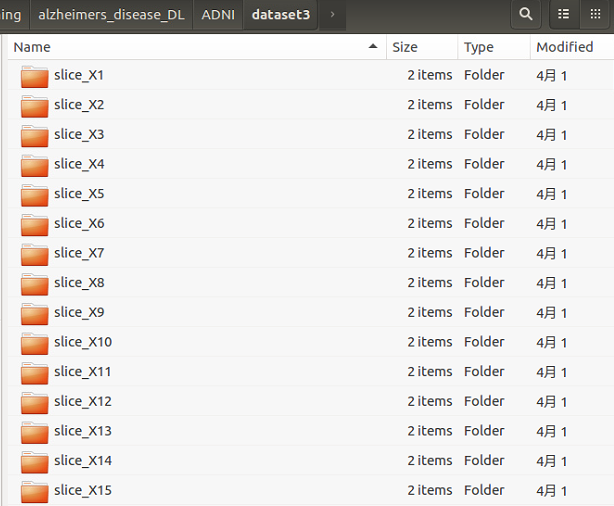
2)方法:將每個位置的切片圖單獨儲存在一個目錄下,總共得到387個目錄,每個目錄下有429張(AD=199張,NC=230張)切片資料;按照8:2的比例劃分 train set 和 validation set;然後分別使用AlexNet進行訓練,記錄 best_val_acc作為評判依據;
處理後的資料形式如下所示:
以 slice_X10 為例:
1)該目錄下有2個子目錄,分別為 train 和 validation;
2)train 和 validation 目錄下也分別有2個子目錄對應2個類別:AD 和 NC;
3)樣本比例:train(AD:NC=159:184);validation(AD:NC=40:46)
原始碼:
step1:注意該指令碼所在的目錄
#!/usr/bin/python # -*- coding: utf-8 -*- import os import re import time import datetime import shutil root_path = "/home/reserch/documents/deeplearning/alzheimers_disease_DL/ADNI" def specified_subject_move_to_fold(slice_path_txt_list, target_path, label): slice_index = 1 subject_num = 0 # print(slice_path_txt_list) # print(target_path) with open(slice_path_txt_list,"r") as slice_txt_path_list: for slice_txt_path in slice_txt_path_list: # slice_txt_path = slice_txt_path_list.readline() slice_txt_path = slice_txt_path.replace("\n", "") slice_txt_path = slice_txt_path.replace("\r", "") slice_txt_path = slice_txt_path.replace("\\", "/") subject_num = subject_num + 1 try: subject_id = slice_txt_path.split("/")[4] # print("subject_id = {}".format(subject_id)) except: subject_id = "" print("...xx...") # print(slice_txt_path) entropy_value_txt_name = "entropy_value_" + label + "_gray_matter_Slices.txt" slice_txt = os.path.join(slice_txt_path, entropy_value_txt_name) # print(slice_txt) with open(slice_txt, "r") as slice_path_list: for item_slice in slice_path_list: new_target_path = target_path slice_name = item_slice.split(",")[0] try: if (slice_name.split(".")[1] == "jpg"): slice_postion = slice_name.split(".")[0] slice_path = os.path.join(slice_txt_path, slice_name) if (os.path.exists(slice_path)): # print("slice_path = {}".format(slice_path)) # new_slice_name = "GM" + label + str("%.5d"%slice_index) + "_" + subject_id + ".jpg" new_slice_name = slice_postion + "_" + subject_id + "_" + "GM" + label + ".jpg" # new_slice_name = "GM" + label + "_" + subject_id + ".jpg" new_target_path = os.path.join(new_target_path, slice_postion, label) if not os.path.exists(new_target_path): print("Create dir = {}".format(new_target_path)) os.makedirs(new_target_path) new_name = os.path.join(new_target_path, new_slice_name) slice_index = slice_index + 1 print("copied the image to {}".format(new_name)) shutil.copyfile(slice_path, new_name) except: pass # print("{} not a jpg file.".format(slice_name)) # if(slice_index > 5): # break # except: # print("[error]...") ### subject_num/3 --> 3 including X Y Z print("subject_num = {}".format(subject_num/3)) print("total slice num = {}".format(slice_index)) ### according to AD_gray_matter_Slices_path.txt file, move all slices to a folder (AD_GM_except_entropy_zero) ### new file: all slices in a folder. AD_GM_except_entropy_zero + NC_GM_except_entropy_zero if __name__=="__main__": dataset = 'dataset3' slice_path_txt_list = './AD_NC_ALL_SLICE/NC_gray_matter_Slices_ALL/NC_gray_matter_Slices_path.txt' target_path = os.path.join(root_path, dataset) label = 'NC' specified_subject_move_to_fold(slice_path_txt_list, target_path, label) slice_path_txt_list = './AD_NC_ALL_SLICE/AD_gray_matter_Slices_ALL/AD_gray_matter_Slices_path.txt' target_path = os.path.join(root_path, dataset) label = 'AD' specified_subject_move_to_fold(slice_path_txt_list, target_path, label)
step2:
#!/usr/bin/python
# -*- coding: utf-8 -*-
import os
import re
import time
import datetime
import shutil
import random
from hcq_lib import *
train_percentage = 0.8
val_percentage = 0.2
test_percentage = 0.1
len_slice_list_CascadeCNNs_AD = 199 ## 199 + 230
len_slice_list_CascadeCNNs_NC = 230 ## 199 + 230
rondom_list_AD = random.sample(range(0, len_slice_list_CascadeCNNs_AD), len_slice_list_CascadeCNNs_AD)
rondom_list_NC = random.sample(range(0, len_slice_list_CascadeCNNs_NC), len_slice_list_CascadeCNNs_NC)
path_backup_random_list = "/home/reserch/documents/deeplearning/alzheimers_disease_DL/ADNI/backup_random_list/rondom_list.txt"
# hcq_backup_txt_rename(path_backup_random_list)
hcq_write(path_backup_random_list, True, True, rondom_list_AD)
hcq_write(path_backup_random_list, True, True, rondom_list_NC)
def get_slice_train_val_test(root_path, slice_folder_path):
## root_path = "/home/reserch/documents/deeplearning/alzheimers_disease_DL/ADNI/dataset3"
## slice_folder_path = slice_X44
## train_target_path = /home/reserch/documents/deeplearning/alzheimers_disease_DL/ADNI/dataset/slice_X44/train
train_target_path = os.path.join(root_path, slice_folder_path, "train")
val_target_path = os.path.join(root_path, slice_folder_path, "validation")
### get all silce through its path
slice_list_AD = os.listdir(os.path.join(root_path, slice_folder_path, "AD"))
slice_list_NC = os.listdir(os.path.join(root_path, slice_folder_path, "NC"))
# len_slice_list_AD = len(slice_list_AD)
# len_slice_list_NC = len(slice_list_NC)
### set the number of train, val, test
# train_num = int(train_percentage * len_slice_list)
# val_num = len_slice_list - train_num
# print("=====")
# print("total_num = {}".format(len_slice_list))
# print("train_num = {}".format(train_num))
# print("val_num = {}".format(val_num))
### create a rondom list without repetition
# rondom_list = random.sample(range(0, len_slice_list), len_slice_list)
# print(rondom_list)
hcq_create_dir(os.path.join(train_target_path, "AD"))
hcq_create_dir(os.path.join(train_target_path, "NC"))
hcq_create_dir(os.path.join(val_target_path, "AD"))
hcq_create_dir(os.path.join(val_target_path, "NC"))
### create txt file to store the index of train, val, test
# train: [0, train_num-1]
num_train_AD = 0
num_train_NC = 0
num_val_AD = 0
num_val_NC = 0
### AD
for i in range(len_slice_list_CascadeCNNs_AD):
slice_index = rondom_list_AD[i]
label = ((slice_list_AD[slice_index].split("_")[3]).split(".")[0])[2:4]
old_path = os.path.join(os.path.join(root_path, slice_folder_path), label, slice_list_AD[slice_index])
if(num_train_AD < int(len_slice_list_CascadeCNNs_AD*train_percentage)):
num_train_AD += 1
new_path = os.path.join(train_target_path, label, slice_list_AD[slice_index])
else:
num_val_AD += 1
new_path = os.path.join(val_target_path, label, slice_list_AD[slice_index])
shutil.copyfile(old_path, new_path)
os.remove(old_path)
# print("===")
# print(old_path)
# print(new_path)
### NC
# # val: [train_num, train_num + val_num - 1]
for i in range(len_slice_list_CascadeCNNs_NC):
slice_index = rondom_list_NC[i]
label = ((slice_list_NC[slice_index].split("_")[3]).split(".")[0])[2:4]
old_path = os.path.join(os.path.join(root_path, slice_folder_path), label, slice_list_NC[slice_index])
if(num_train_NC < int(len_slice_list_CascadeCNNs_NC*train_percentage)):
num_train_NC += 1
new_path = os.path.join(train_target_path, label, slice_list_NC[slice_index])
else:
num_val_NC += 1
new_path = os.path.join(val_target_path, label, slice_list_NC[slice_index])
shutil.copyfile(old_path, new_path)
os.remove(old_path)
# print("===")
# print(old_path)
# print(new_path)
print("num_train_AD = {}".format(num_train_AD))
print("num_train_NC = {}".format(num_train_NC))
print("num_val_AD = {}".format(num_val_AD))
print("num_val_NC = {}".format(num_val_NC))
### delete empty folder: AD, NC
hcq_rmdir(os.path.join(root_path, slice_folder_path, "AD"))
hcq_rmdir(os.path.join(root_path, slice_folder_path, "NC"))
if __name__=="__main__":
root_path = "/home/reserch/documents/deeplearning/alzheimers_disease_DL/ADNI/dataset3"
slice_folder_list = os.listdir(root_path)
num = 0
for slice_folder_path in slice_folder_list:
num += 1
# print("===")
# print(num)
get_slice_train_val_test(root_path, slice_folder_path)
# if(num>0):
# break