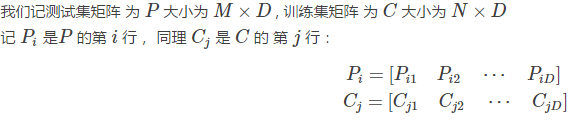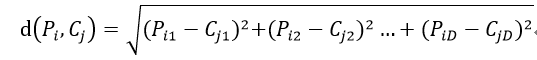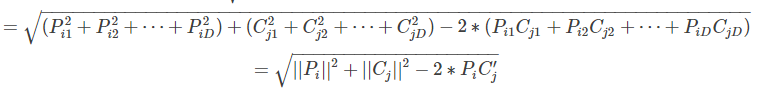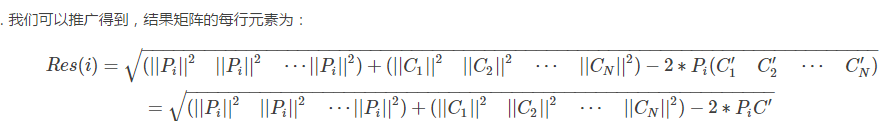執行cs231n課程中Assignment1中的示例程式碼
因為我用的版本是python3.6.0,而示例程式碼似乎是python2版本的,於是遇到一些問題。
示例程式碼是ipynb格式的,開啟方式:cmd下執行ipython notebook, 在瀏覽器中開啟網頁(如下),可以點選Upload按鈕選擇要開啟的檔案。
但是程式碼是一行一行的,怎麼在spyder裡面直接執行呢?
如圖2點選儲存為python格式的就可以在spyder裡面打開了。
一執行發現報錯了,原來資料集沒有下載:
回頭看作業頁面的說明,原來說了要怎麼下載的:
但是這個是Shell命令。(windows10上可以啟用linux子系統)開啟bash,輸入命令,開始下載CIFAR-10 dataset
執行程式碼的時候報了許多錯,其中之一是pickle.load反序列化,
File "E:\assignment1\cs231n\data_utils.py", line 9, in load_CIFAR_batch
datadict = pickle.load(f)
UnicodeDecodeError: 'ascii' codec can't decode byte 0x8b in position 6: ordinal not in range(128)查到別人的解答,點選開啟連結
with open(filename, 'rb') as f: datadict = pickle.load(f,encoding='iso-8859-1')
我們需要告訴pickle:how to convert python bytestring data to Python 3 strings,The default is to try and decode all string data as ASCII
填補程式碼說明:
- 交叉驗證選擇最佳的K值
# In[ ]: num_folds = 5 k_choices = [1, 3, 5, 8, 10, 12, 15, 20, 50, 100] X_train_folds = [] y_train_folds = [] ################################################################################ # TODO: # # Split up the training data into folds. After splitting, X_train_folds and # # y_train_folds should each be lists of length num_folds, where # # y_train_folds[i] is the label vector for the points in X_train_folds[i]. # # Hint: Look up the numpy array_split function. # ################################################################################ X_train_folds=np.array_split(X_train,num_folds) y_train_folds=np.array_split(y_train,num_folds) ################################################################################ # END OF YOUR CODE # ################################################################################ # A dictionary holding the accuracies for different values of k that we find # when running cross-validation. After running cross-validation, # k_to_accuracies[k] should be a list of length num_folds giving the different # accuracy values that we found when using that value of k. k_to_accuracies = {} ################################################################################ # TODO: # # Perform k-fold cross validation to find the best value of k. For each # # possible value of k, run the k-nearest-neighbor algorithm num_folds times, # # where in each case you use all but one of the folds as training data and the # # last fold as a validation set. Store the accuracies for all fold and all # # values of k in the k_to_accuracies dictionary. # ################################################################################ for k in k_choices: k_to_accuracies[k] = np.zeros(num_folds) for i in range(num_folds): Xtr = np.array(X_train_folds[:i] + X_train_folds[i+1:])#把訓練集中的一塊劃為驗證集 ytr = np.array(y_train_folds[:i] + y_train_folds[i+1:]) Xte = np.array(X_train_folds[i]) yte = np.array(y_train_folds[i]) Xtr = np.reshape(Xtr, (X_train.shape[0]/ 5*4, -1)) ytr = np.reshape(ytr, (y_train.shape[0]/ 5*4, -1)) Xte = np.reshape(Xte, (X_train.shape[0] / 5, -1)) yte = np.reshape(yte, (y_train.shape[0] / 5, -1)) classifier.train(Xtr, ytr) yte_pred = classifier.predict(Xte, k) yte_pred = np.reshape(yte_pred, (yte_pred.shape[0], -1)) num_correct = np.sum(yte_pred == yte) accuracy = float(num_correct) / len(yte) k_to_accuracies[k][i] = accuracy ################################################################################ # END OF YOUR CODE # ################################################################################ # Print out the computed accuracies for k in sorted(k_to_accuracies): for accuracy in k_to_accuracies[k]: print ('k = %d, accuracy = %f' % (k, accuracy))
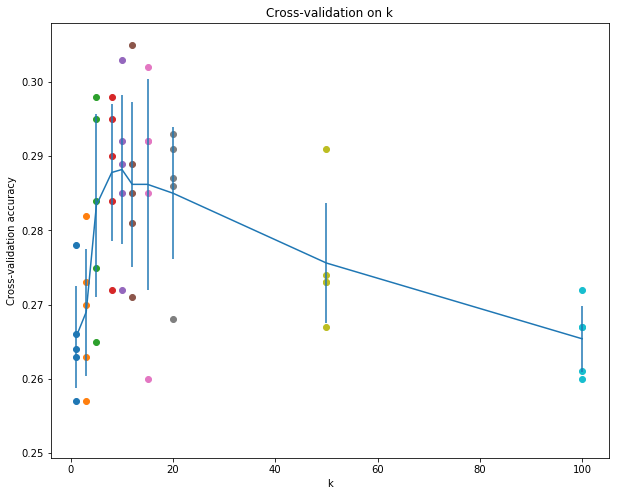
這裡使用的是交叉驗證(cross-validation)去獲取knn中的超引數K的最佳值,其基本思想是,將訓練集劃分為num_folds個塊,迴圈地把其中的一塊作為驗證集,計算不同K值下載所有遍歷的驗證集上的準確率(num_folds個結果)的平均值作為該K值在訓練集上的準確率,然後選擇這個準確率最高的K值作為KNN中的K。
在這個試驗中,num_folds的值為5,也就是把訓練集分作了5塊,這個操作是使用
np.array_split(X_train,num_folds)然後再使用一個迴圈,其次數為num_folds次,每一次迴圈裡選擇其中一塊作為驗證集,然後把剩下的作為訓練集,呼叫train函式訓練(KNN並不訓練,只是儲存了訓練資料),然後使用predict得到驗證集的預測標籤,判斷預測標籤和實際標籤對應的正確率,作為該K值下該塊驗證集對應的準確率,執行結果如下:
k = 1, accuracy = 0.263000
k = 1, accuracy = 0.257000
k = 1, accuracy = 0.264000
k = 1, accuracy = 0.278000
k = 1, accuracy = 0.266000
k = 3, accuracy = 0.257000
k = 3, accuracy = 0.263000
k = 3, accuracy = 0.273000
k = 3, accuracy = 0.282000
k = 3, accuracy = 0.270000
k = 5, accuracy = 0.265000
k = 5, accuracy = 0.275000
k = 5, accuracy = 0.295000
k = 5, accuracy = 0.298000
k = 5, accuracy = 0.284000
k = 8, accuracy = 0.272000
k = 8, accuracy = 0.295000
k = 8, accuracy = 0.284000
k = 8, accuracy = 0.298000
k = 8, accuracy = 0.290000
k = 10, accuracy = 0.272000
k = 10, accuracy = 0.303000
k = 10, accuracy = 0.289000
k = 10, accuracy = 0.292000
k = 10, accuracy = 0.285000
k = 12, accuracy = 0.271000
k = 12, accuracy = 0.305000
k = 12, accuracy = 0.285000
k = 12, accuracy = 0.289000
k = 12, accuracy = 0.281000
k = 15, accuracy = 0.260000
k = 15, accuracy = 0.302000
k = 15, accuracy = 0.292000
k = 15, accuracy = 0.292000
k = 15, accuracy = 0.285000
k = 20, accuracy = 0.268000
k = 20, accuracy = 0.293000
k = 20, accuracy = 0.291000
k = 20, accuracy = 0.287000
k = 20, accuracy = 0.286000
k = 50, accuracy = 0.273000
k = 50, accuracy = 0.291000
k = 50, accuracy = 0.274000
k = 50, accuracy = 0.267000
k = 50, accuracy = 0.273000
k = 100, accuracy = 0.261000
k = 100, accuracy = 0.272000
k = 100, accuracy = 0.267000
k = 100, accuracy = 0.260000
k = 100, accuracy = 0.267000準確率也都不算高,28%左右。
將每個K值對應的5個驗證集上的準確率取平均值,繪製折線圖如下:
可以看到,第5條豎線對應的均值達到了折線的頂峰,在K_choice裡面第五個K值對應的是K=10,因此best_k的值應該改為10
# Based on the cross-validation results above, choose the best value for k,
# retrain the classifier using all the training data, and test it on the test
# data. You should be able to get above 28% accuracy on the test data.
best_k = 10
classifier = KNearestNeighbor()
classifier.train(X_train, y_train)
y_test_pred = classifier.predict(X_test, k=best_k)
# Compute and display the accuracy
num_correct = np.sum(y_test_pred == y_test)
accuracy = float(num_correct) / num_test
print ('Got %d / %d correct => accuracy: %f' % (num_correct, num_test, accuracy))提示說最後的準確率應該比28%大,如果best_k取1時候,執行結果的準確率是27%左右,當把K改為10以後,準確率是:
Got 144 / 500 correct => accuracy: 0.288000而沒有使用交叉驗證時候,指定K=1和K=5時候的準確率分別為27%和29%(這裡沒弄明白為什麼交叉驗證選出的K=10在測試集上的表現不如動手指定的k=5,可能因為選用的訓練集並不是全部訓練集)
Got 137 / 500 correct => accuracy: 0.274000 #k=1
Got 145 / 500 correct => accuracy: 0.290000 #k=52.使用歐式距離計算測試資料和訓練資料的距離矩陣,三種方法,一種使用兩層迴圈,二種使用一層迴圈,最後一種需要用到更多的數學知識和向量化處理,不使用迴圈進行計算:
def compute_distances_two_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a nested loop over both the training data and the
test data.
Inputs:
- X: A numpy array of shape (num_test, D) containing test data.
Returns:
- dists: A numpy array of shape (num_test, num_train) where dists[i, j]
is the Euclidean distance between the ith test point and the jth training
point.
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in range(num_test):
for j in range(num_train):
#####################################################################
# TODO: #
# Compute the l2 distance between the ith test point and the jth #
# training point, and store the result in dists[i, j]. You should #
# not use a loop over dimension. #
#####################################################################
dists[i,j]=np.sqrt(np.sum(np.square(self.X_train[j,:]-X[i,:])))
#####################################################################
# END OF YOUR CODE #
#####################################################################
return dists
def compute_distances_one_loop(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using a single loop over the test data.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
for i in range(num_test):
#######################################################################
# TODO: #
# Compute the l2 distance between the ith test point and all training #
# points, and store the result in dists[i, :]. #
#######################################################################
dists[i,:] = np.sqrt(np.sum(np.square(self.X_train-X[i,:]),axis = 1))
#np.square是針對每個元素的平方方法
#######################################################################
# END OF YOUR CODE #
#######################################################################
return dists
def compute_distances_no_loops(self, X):
"""
Compute the distance between each test point in X and each training point
in self.X_train using no explicit loops.
Input / Output: Same as compute_distances_two_loops
"""
num_test = X.shape[0]
num_train = self.X_train.shape[0]
dists = np.zeros((num_test, num_train))
#########################################################################
# TODO: #
# Compute the l2 distance between all test points and all training #
# points without using any explicit loops, and store the result in #
# dists. #
# #
# You should implement this function using only basic array operations; #
# in particular you should not use functions from scipy. #
# #
# HINT: Try to formulate the l2 distance using matrix multiplication #
# and two broadcast sums. #
#########################################################################
dists = np.multiply(np.dot(X,self.X_train.T),-2)
sq1 = np.sum(np.square(X),axis=1,keepdims = True)
sq2 = np.sum(np.square(self.X_train),axis=1)
dists = np.add(dists,sq1)
dists = np.add(dists,sq2)
dists = np.sqrt(dists)
#########################################################################
# END OF YOUR CODE #
#########################################################################
return dists第一種執行的時間效率最低的是使用兩層迴圈,計算每個測試向量和每個訓練向量的向量差,然後使用np.square()計算求得每個元素的平方(也可以使用點乘的方式:np.dot(X[i] - self.X_train[j], X[i] - self.X_train[j])),使用np.sum()計算元素平方的和,最後開方求得的就是該測試向量和訓練向量的歐式距離,所有遍歷下來需要計算500*5000次,也就是測試資料集的行數乘以訓練資料集的行數。
5000行的3072維訓練資料,500行的3072維測試資料,計算的時間花銷是57秒左右:
Two loop version took 57.858908 seconds第二種方法,使用一層迴圈,遍歷每個測試向量,直接計算每個測試向量(1*3072)和所有訓練向量(5000*3072)的差,得到一個5000*3072的矩陣,然後使用np.square(X)計算每個元素的平方,再使用np.sum(np.square(self.X_train-X[i,:]),axis = 1)計算每行所有列的元素平方的和,再開方得到一個5000*1的向量,以橫向量形式儲存在結果陣列的一行 中,表示的是該測試向量和所有訓練向量的歐式距離,最後得到的就是500*5000的一個結果,也就是測試資料集的行數乘以訓練資料集的行數。
使用一層迴圈的時間開銷是使用兩層迴圈的兩倍,大約106秒:
One loop version took 106.198080 seconds最後一種方式不使用迴圈,但是需要用到一點推理歸納:
我們先來計算一下 Pi 和 Cj 之間的距離
因此,結果矩陣的表示形式為:
換成python程式碼就是:
dists = np.multiply(np.dot(X,self.X_train.T),-2) #維度是(500,5000)
sq1 = np.sum(np.square(X),axis=1,keepdims = True) #維度是(500,1)
sq2 = np.sum(np.square(self.X_train),axis=1) #維度是(5000,)沒有保持維度,也不能保持維度
dists = np.add(dists,sq1) #維度是(500,5000)
dists = np.add(dists,sq2)
dists = np.sqrt(dists)值得注意的是程式碼中先計算了最後一部分2PC',此時資料維度是500*5000,然後計算測試資料和訓練資料的元素平方,但是訓練資料不能保持維度,最後使用np.add函式對兩個向量和前面的矩陣進行加法,維度不變,依然是500*5000.