使用python做遺傳演算法與基於遺傳演算法的多目標演算法
阿新 • • 發佈:2019-02-08
- 遺傳演算法
建立GeneticAlgorithm.py
import numpy as np
from GAIndividual import GAIndividual
import random
import copy
import matplotlib.pyplot as plt
class GeneticAlgorithm:
'''
The class for genetic algorithm
'''
def __init__(self, sizepop, vardim, bound, MAXGEN, params):
'''
sizepop: population sizepop 種群數量 60
vardim: dimension of variables 變數維度 25
bound: boundaries of variables 變數的邊界 -600 600
MAXGEN: termination condition 終止條件 1000
param: algorithm required parameters, it is a list which is consisting of crossover rate, mutation rate, alpha
演算法所需的引數,它是由交叉率,變異率,alpha組成的列表
0.9, 0.1, 0.5
''' 建立GAIndividual.py
import numpy as np
import ObjFunction
#個體的遺傳演算法
class GAIndividual:
'''
individual of genetic algorithm
個體的遺傳演算法
'''
def __init__(self, vardim, bound):
'''
vardim: dimension of variables 維度變數
bound: boundaries of variables 變數的邊界
'''
self.vardim = vardim
self.bound = bound
self.fitness = 0.
def generate(self):
'''
generate a random chromsome for genetic algorithm
為遺傳演算法生成一個隨機染色體
'''
len = self.vardim
rnd = np.random.random(size=len)
self.chrom = np.zeros(len)
for i in range(0, len):
self.chrom[i] = self.bound[0, i] + \
(self.bound[1, i] - self.bound[0, i]) * rnd[i]
def calculateFitness(self):
'''
calculate the fitness of the chromsome
計算染色體的適應性
'''
self.fitness = ObjFunction.GrieFunc(
self.vardim, self.chrom, self.bound)三建立ObjFunction.py
import math
#目標函式
def GrieFunc(vardim, x, bound):
"""
Griewangk function
經典函式girewangk
"""
s1 = 0.
s2 = 1.
for i in range(1, vardim + 1):
s1 = s1 + x[i - 1] ** 2
s2 = s2 * math.cos(x[i - 1] / math.sqrt(i))
y = (1. / 4000.) * s1 - s2 + 1
y = 1. / (1. + y)
return y
#非凸優化函式
def RastFunc(vardim, x, bound):
"""
Rastrigin function
在數學優化中,Rastrigin函式是一個非凸函式,用作優化演算法的效能測試問題。這是一個非線性多模態函式的典型例子。它最初由Rastrigin [1]提出作為二維函式,並已被Mühlenbein等人推廣。[2]尋找這個函式的最小值是一個相當困難的問題,因為它有很大的搜尋空間和大量的區域性最小值。
在一個n維域上,它被定義為:
{\ displaystyle f(\ mathbf {x})= An + \ sum _ {i = 1} ^ {n} \ left [x_ {i} ^ {2} -A \ cos(2 \ pi x_ {i})\對]} f(\ mathbf {x})= An + \ sum _ {i = 1} ^ {n} \ left [x_ {i} ^ {2} -A \ cos(2 \ pi x_ {i})\ right]
"""
s = 10 * 25
for i in range(1, vardim + 1):
s = s + x[i - 1] ** 2 - 10 * math.cos(2 * math.pi * x[i - 1])
return s基於遺傳演算法的多目標演算法
#Importing required modules
import math
import random
import matplotlib.pyplot as plt
def function1(x):
value = -x**2
return value
def function2(x):
value = -(x-2)**2
return value
#Function to find index of list
#函式查詢列表的索引
def index_of(a,list):
for i in range(0,len(list)):
if list[i] == a:
return i
return -1
#Function to sort by values 函式根據值排序
def sort_by_values(list1, values):
sorted_list = []
while(len(sorted_list)!=len(list1)):
if index_of(min(values),values) in list1:
sorted_list.append(index_of(min(values),values))
values[index_of(min(values),values)] = math.inf
return sorted_list
#Function to carry out NSGA-II's fast non dominated sort
#函式執行NSGA-II的快速非支配排序
"""基於序列和擁擠距離"""
def fast_non_dominated_sort(values1, values2):
S=[[] for i in range(0,len(values1))]
front = [[]]
n=[0 for i in range(0,len(values1))]
rank = [0 for i in range(0, len(values1))]
for p in range(0,len(values1)):
S[p]=[]
n[p]=0
for q in range(0, len(values1)):
#p > q
if (values1[p] > values1[q] and values2[p] > values2[q]) or (values1[p] >= values1[q] and values2[p] > values2[q]) or (values1[p] > values1[q] and values2[p] >= values2[q]):
if q not in S[p]:
S[p].append(q)
elif (values1[q] > values1[p] and values2[q] > values2[p]) or (values1[q] >= values1[p] and values2[q] > values2[p]) or (values1[q] > values1[p] and values2[q] >= values2[p]):
n[p] = n[p] + 1
if n[p]==0:
rank[p] = 0
if p not in front[0]:
front[0].append(p)
i = 0
while(front[i] != []):
Q=[]
for p in front[i]:
for q in S[p]:
n[q] =n[q] - 1
if( n[q]==0):
rank[q]=i+1
if q not in Q:
Q.append(q)
i = i+1
front.append(Q)
del front[len(front)-1]
return front
#Function to calculate crowding distance
#計算擁擠距離的函式
def crowding_distance(values1, values2, front):
distance = [0 for i in range(0,len(front))]
sorted1 = sort_by_values(front, values1[:])
sorted2 = sort_by_values(front, values2[:])
distance[0] = 4444444444444444
distance[len(front) - 1] = 4444444444444444
for k in range(1,len(front)-1):
distance[k] = distance[k]+ (values1[sorted1[k+1]] - values2[sorted1[k-1]])/(max(values1)-min(values1))
for k in range(1,len(front)-1):
distance[k] = distance[k]+ (values1[sorted2[k+1]] - values2[sorted2[k-1]])/(max(values2)-min(values2))
return distance
#Function to carry out the crossover
#函式進行交叉
def crossover(a,b):
r=random.random()
if r>0.5:
return mutation((a+b)/2)
else:
return mutation((a-b)/2)
#Function to carry out the mutation operator
#函式進行變異操作
def mutation(solution):
mutation_prob = random.random()
if mutation_prob <1:
solution = min_x+(max_x-min_x)*random.random()
return solution
#Main program starts here
pop_size = 20
max_gen = 921
#Initialization
min_x=-55
max_x=55
solution=[min_x+(max_x-min_x)*random.random() for i in range(0,pop_size)]
gen_no=0
while(gen_no<max_gen):
function1_values = [function1(solution[i])for i in range(0,pop_size)]
function2_values = [function2(solution[i])for i in range(0,pop_size)]
non_dominated_sorted_solution = fast_non_dominated_sort(function1_values[:],function2_values[:])
print("The best front for Generation number ",gen_no, " is")
for valuez in non_dominated_sorted_solution[0]:
print(round(solution[valuez],3),end=" ")
print("\n")
crowding_distance_values=[]
for i in range(0,len(non_dominated_sorted_solution)):
crowding_distance_values.append(crowding_distance(function1_values[:],function2_values[:],non_dominated_sorted_solution[i][:]))
solution2 = solution[:]
#Generating offsprings
while(len(solution2)!=2*pop_size):
a1 = random.randint(0,pop_size-1)
b1 = random.randint(0,pop_size-1)
solution2.append(crossover(solution[a1],solution[b1]))
function1_values2 = [function1(solution2[i])for i in range(0,2*pop_size)]
function2_values2 = [function2(solution2[i])for i in range(0,2*pop_size)]
non_dominated_sorted_solution2 = fast_non_dominated_sort(function1_values2[:],function2_values2[:])
crowding_distance_values2=[]
for i in range(0,len(non_dominated_sorted_solution2)):
crowding_distance_values2.append(crowding_distance(function1_values2[:],function2_values2[:],non_dominated_sorted_solution2[i][:]))
new_solution= []
for i in range(0,len(non_dominated_sorted_solution2)):
non_dominated_sorted_solution2_1 = [index_of(non_dominated_sorted_solution2[i][j],non_dominated_sorted_solution2[i] ) for j in range(0,len(non_dominated_sorted_solution2[i]))]
front22 = sort_by_values(non_dominated_sorted_solution2_1[:], crowding_distance_values2[i][:])
front = [non_dominated_sorted_solution2[i][front22[j]] for j in range(0,len(non_dominated_sorted_solution2[i]))]
front.reverse()
for value in front:
new_solution.append(value)
if(len(new_solution)==pop_size):
break
if (len(new_solution) == pop_size):
break
solution = [solution2[i] for i in new_solution]
gen_no = gen_no + 1
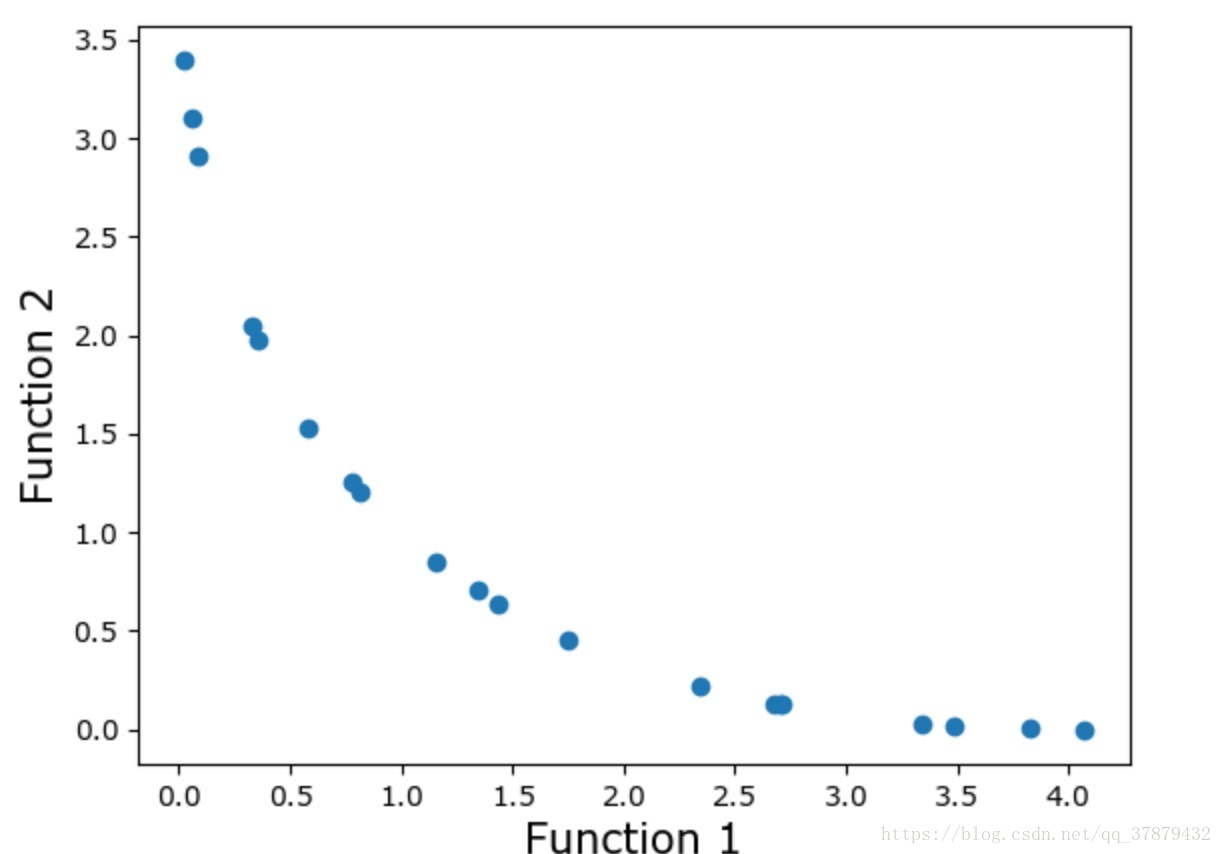
#Lets plot the final front now
function1 = [i * -1 for i in function1_values]
function2 = [j * -1 for j in function2_values]
plt.xlabel('Function 1', fontsize=15)
plt.ylabel('Function 2', fontsize=15)
plt.scatter(function1, function2)
plt.show()